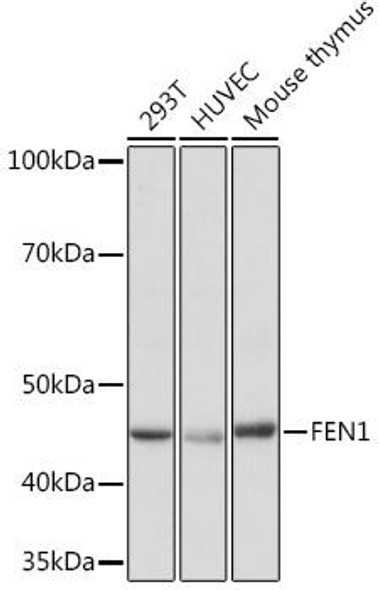
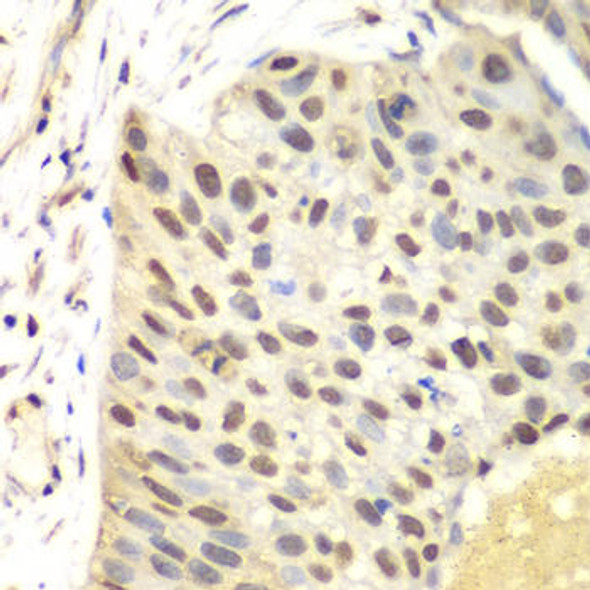
Description
| Antibody Name: | FEN1 Antibody (PACO63543) |
| Antibody SKU: | PACO63543 |
| Size: | 50ul |
| Host Species: | Rabbit |
| Tested Applications: | ELISA, WB |
| Recommended Dilutions: | ELISA:1:2000-1:10000, WB:1:1000-1:5000 |
| Species Reactivity: | Arabidopsis thaliana |
| Immunogen: | Recombinant Arabidopsis thaliana Flap endonuclease 1 protein (85-383AA) |
| Form: | Liquid |
| Storage Buffer: | Preservative: 0.03% Proclin 300 Constituents: 50% Glycerol, 0.01M PBS, pH 7.4 |
| Purification Method: | >95%, Protein G purified |
| Clonality: | Polyclonal |
| Isotype: | IgG |
| Conjugate: | Non-conjugated |
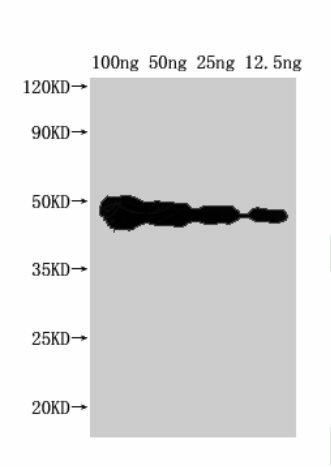 | Western Blot. Positive WB detected in Recombinant protein. All lanes: FEN1 antibody at 1:2000. Secondary. Goat polyclonal to rabbit IgG at 1/50000 dilution. Predicted band size: 37 kDa. Observed band size: 45 kDa. |
| Background: | Structure-specific nuclease with 5'-flap endonuclease and 5'-3' exonuclease activities involved in DNA replication and repair. During DNA replication, cleaves the 5'-overhanging flap structure that is generated by displacement synthesis when DNA polymerase encounters the 5'-end of a downstream Okazaki fragment. It enters the flap from the 5'-end and then tracks to cleave the flap base, leaving a nick for ligation. Also involved in the long patch base excision repair (LP-BER) pathway, by cleaving within the apurinicapyrimidinic (AP) site-terminated flap. Acts as a genome stabilization factor that prevents flaps from equilibrating into structurs that lead to duplications and deletions. Also possesses 5'-3' exonuclease activity on nicked or gapped double-stranded DNA, and exhibits RNase H activity. Also involved in replication and repair of rDNA and in repairing mitochondrial DNA. |
| Synonyms: | Flap endonuclease 1, FEN-1, Flap structure-specific endonuclease 1, FEN1 |
| UniProt Protein Function: | Structure-specific nuclease with 5'-flap endonuclease and 5'-3' exonuclease activities involved in DNA replication and repair. During DNA replication, cleaves the 5'-overhanging flap structure that is generated by displacement synthesis when DNA polymerase encounters the 5'-end of a downstream Okazaki fragment. It enters the flap from the 5'-end and then tracks to cleave the flap base, leaving a nick for ligation. Also involved in the long patch base excision repair (LP-BER) pathway, by cleaving within the apurinic/apyrimidinic (AP) site-terminated flap. Acts as a genome stabilization factor that prevents flaps from equilibrating into structurs that lead to duplications and deletions. Also possesses 5'-3' exonuclease activity on nicked or gapped double-stranded DNA, and exhibits RNase H activity. Also involved in replication and repair of rDNA and in repairing mitochondrial DNA. |
| UniProt Protein Details: | |
| NCBI Summary: | |
| UniProt Code: | O65251 |
| NCBI GenInfo Identifier: | 238481386 |
| NCBI Gene ID: | 832721 |
| NCBI Accession: | NP_001154740.1 |
| UniProt Secondary Accession: | O65251 |
| UniProt Related Accession: | O65251 |
| Molecular Weight: | 42,797 Da |
| NCBI Full Name: | 5'-3' exonuclease family protein |
| NCBI Synonym Full Names: | |
| NCBI Official Symbol: | AT5G26680 |
| NCBI Official Synonym Symbols: | |
| NCBI Protein Information: | 5'-3' exonuclease family protein |
| UniProt Protein Name: | Flap endonuclease 1 |
| UniProt Synonym Protein Names: | Flap structure-specific endonuclease 1 |
| Protein Family: | |
| UniProt Gene Name: | FEN1 |
| UniProt Entry Name: |