Epigenetics & Nuclear Signaling Antibodies 5
Anti-Phospho-TP53BP1-T543 pAb Antibody (CABP0861)
- SKU:
- CABP0861
- Product Type:
- Antibody
- Applications:
- WB
- Reactivity:
- Human
- Host Species:
- Rabbit
- Isotype:
- IgG
- Research Area:
- Epigenetics and Nuclear Signaling
Description
| Antibody Name: | Anti-Phospho-TP53BP1-T543 Antibody |
| Antibody SKU: | CABP0861 |
| Antibody Size: | 20uL, 50uL, 100uL |
| Application: | WB |
| Reactivity: | Human |
| Host Species: | Rabbit |
| Immunogen: | A synthetic phosphorylated peptide around T543 of human TP53BP1 (NP_001135451.1). |
| Application: | WB |
| Recommended Dilution: | WB 1:500 - 1:2000 |
| Reactivity: | Human |
| Positive Samples: |
| Immunogen: | A synthetic phosphorylated peptide around T543 of human TP53BP1 (NP_001135451.1). |
| Purification Method: | Affinity purification |
| Storage Buffer: | Store at -20°C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | IDED G |
| Gene ID: | 7158 |
| Uniprot: | Q12888 |
| Cellular Location: | Chromosome, Nucleus, centromere, kinetochore |
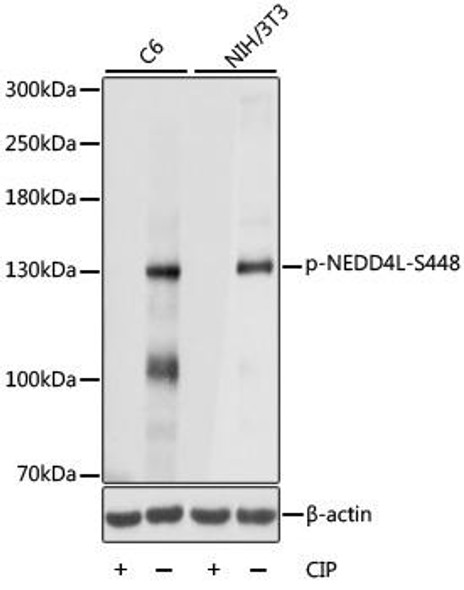
| Calculated MW: | 213kDa/214kDa |
| Observed MW: | Refer to figures |
| Synonyms: | TP53BP1, 53BP1, TDRD30, TP53, p202, p53BP1 |
| Background: | Double-strand break (DSB) repair protein involved in response to DNA damage, telomere dynamics and class-switch recombination (CSR) during antibody genesis (PubMed:12364621, PubMed:22553214, PubMed:23333306, PubMed:17190600, PubMed:21144835, PubMed:28241136). Plays a key role in the repair of double-strand DNA breaks (DSBs) in response to DNA damage by promoting non-homologous end joining (NHEJ)-mediated repair of DSBs and specifically counteracting the function of the homologous recombination (HR) repair protein BRCA1 (PubMed:22553214, PubMed:23727112, PubMed:23333306). In response to DSBs, phosphorylation by ATM promotes interaction with RIF1 and dissociation from NUDT16L1/TIRR, leading to recruitment to DSBs sites (PubMed:28241136). Recruited to DSBs sites by recognizing and binding histone H2A monoubiquitinated at 'Lys-15' (H2AK15Ub) and histone H4 dimethylated at 'Lys-20' (H4K20me2), two histone marks that are present at DSBs sites (PubMed:23760478, PubMed:28241136, PubMed:17190600). Required for immunoglobulin class-switch recombination (CSR) during antibody genesis, a process that involves the generation of DNA DSBs (PubMed:23345425). Participates to the repair and the orientation of the broken DNA ends during CSR (By similarity). In contrast, it is not required for classic NHEJ and V(D)J recombination (By similarity). Promotes NHEJ of dysfunctional telomeres via interaction with PAXIP1 (PubMed:23727112). |
| UniProt Protein Function: | 53BP1: a DNA damage checkpoint protein that interacts with HDAC4 protein to mediate the DNA damage response. Inhibits E2F1-mediated apoptosis in prostate cancer cells. Recruited to double strand breaks via the binding of its Tudor domain to dimethylated H3 K20. Asymmetrically dimethylated on Arg residues by PRMT1. Methylation is required for DNA binding. Binds to the central domain of p53. Interacts with Artemis. Interacts with histone H2AFX and this requires phosphorylation of H2AFX on S139. Interacts with histone H4 that has been dimethylated at K20. Has low affinity for histone H4 containing monomethylated K20. Does not bind histone H4 containing unmethylated or trimethylated K20. Has low affinity for histone H3 that has been dimethylated on K79. Does not bind unmethylated histone H3. Phosphorylated at basal level in the absence of DNA damage. Hyper-phosphorylated in an ATM-dependent manner in response to DNA damage induced by ionizing radiation. Hyper-phosphorylated in an ATR-dependent manner in response to DNA damage induced by UV irradiation. Two alternatively spliced human isoforms have been observed. |
| UniProt Protein Details: | Protein type:DNA repair, damage; Transcription, coactivator/corepressor Chromosomal Location of Human Ortholog: 15q15-q21 Cellular Component: nucleoplasm; chromosome, telomeric region; cytoplasm; replication fork; nucleus Molecular Function:protein binding; p53 binding; damaged DNA binding; telomeric DNA binding; methylated histone residue binding Biological Process: transcription from RNA polymerase II promoter; positive regulation of transcription, DNA-dependent; double-strand break repair; positive regulation of transcription from RNA polymerase II promoter; positive regulation of transcription factor activity; DNA repair; response to DNA damage stimulus; double-strand break repair via homologous recombination |
| UniProt Code: | Q12888 |
| NCBI GenInfo Identifier: | 8928568 |
| NCBI Gene ID: | 7158 |
| NCBI Accession: | Q12888.2 |
| UniProt Secondary Accession: | Q12888,Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4, F8VY86, |
| UniProt Related Accession: | Q12888 |
| Molecular Weight: | 1972 |
| NCBI Full Name: | Tumor suppressor p53-binding protein 1 |
| NCBI Synonym Full Names: | tumor protein p53 binding protein 1 |
| NCBI Official Symbol: | TP53BP1 |
| NCBI Official Synonym Symbols: | p202; 53BP1 |
| NCBI Protein Information: | tumor suppressor p53-binding protein 1; p53BP1; tumor protein 53-binding protein, 1; tumor protein p53-binding protein, 1 |
| UniProt Protein Name: | Tumor suppressor p53-binding protein 1 |
| Protein Family: | Tumor suppressor p53-binding protein |
| UniProt Gene Name: | TP53BP1 |
| UniProt Entry Name: | TP53B_HUMAN |