Rat Signaling ELISA Kits 2
Rat BAX (BCL-2 Associated X Protein) CLIA Kit (RTES00059)
- SKU:
- RTES00059
- Product Type:
- ELISA Kit
- ELISA Type:
- CLIA Kit
- Size:
- 96 Assays
- Sensitivity:
- 18.75pg/mL
- Range:
- 31.25-2000pg/mL
- ELISA Type:
- Sandwich
- Synonyms:
- BCL2L4
- Reactivity:
- Rat
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Cell Death
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Rat |
| Detection method: | Chemiluminescence |
| Detection range: | 31.25-2000 pg/mL |
| Sensitivity: | 18.75 pg/mL |
| Sample volume: | 100µL |
| Sample type: | Serum, plasma and other biological fluids |
| Repeatability: | CV < 15% |
| Specificity: | This kit recognizes Rat BAX in samples. No significant cross-reactivity or interference between Rat BAX and analogues was observed. |
This kit uses Sandwich-CLIA as the method. The micro CLIA plate provided in this kit has been pre-coated with an antibody specific to Rat BAX. Standards or samples are added to the appropriate micro CLIA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Rat BAX and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Rat BAX, biotinylated detection antibody and Avidin-HRP conjugate will appear fluorescence. The Relative light unit (RLU) value is measured spectrophotometrically by the Chemiluminescence immunoassay analyzer. The RLU value is positively associated with the concentration of Rat BAX. The concentration of Rat BAX in the samples can be calculated by comparing the RLU of the samples to the standard curve.
| UniProt Protein Function: | BAX: Accelerates programmed cell death by binding to, and antagonizing the apoptosis repressor BCL2 or its adenovirus homolog E1B 19k protein. Under stress conditions, undergoes a conformation change that causes translocation to the mitochondrion membrane, leading to the release of cytochrome c that then triggers apoptosis. Promotes activation of CASP3, and thereby apoptosis. Homodimer. Forms higher oligomers under stress conditions. Interacts with BCL2L11. Interaction with BCL2L11 promotes BAX oligomerization and association with mitochondrial membranes, with subsequent release of cytochrome c. Forms heterodimers with BCL2, E1B 19K protein, BCL2L1 isoform Bcl-X(L), BCL2L2, MCL1 and A1. Interacts with SH3GLB1 and HN. Interacts with SFN and YWHAZ; the interaction occurs in the cytoplasm. Under stress conditions, JNK-mediated phosphorylation of SFN and YWHAZ, releases BAX to mitochondria. Isoform Sigma interacts with BCL2A1 and BCL2L1 isoform Bcl-X(L). Interacts with RNF144B, which regulates the ubiquitin-dependent stability of BAX. Interacts with CLU under stress conditions that cause a conformation change leading to BAX oligomerization and association with mitochondria. Does not interact with CLU in unstressed cells. Interacts with FAIM2/LFG2. Interacts with human cytomegalovirus/HHV-5 protein vMIA/UL37. Expressed in a wide variety of tissues. Isoform Psi is found in glial tumors. Isoform Alpha is expressed in spleen, breast, ovary, testis, colon and brain, and at low levels in skin and lung. Isoform Sigma is expressed in spleen, breast, ovary, testis, lung, colon, brain and at low levels in skin. Isoform Alpha and isoform Sigma are expressed in pro- myelocytic leukemia, histiocytic lymphoma, Burkitt's lymphoma, T- cell lymphoma, lymphoblastic leukemia, breast adenocarcinoma, ovary adenocarcinoma, prostate carcinoma, prostate adenocarcinoma, lung carcinoma, epidermoid carcinoma, small cell lung carcinoma and colon adenocarcinoma cell lines. Belongs to the Bcl-2 family. 8 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:Tumor suppressor; Membrane protein, integral; Apoptosis; Mitochondrial Cellular Component: cytoplasm; cytosol; endoplasmic reticulum; endoplasmic reticulum membrane; integral to membrane; intracellular; membrane; mitochondrial membrane; mitochondrial outer membrane; mitochondrial permeability transition pore complex; mitochondrion; nuclear envelope; nucleus; perinuclear region of cytoplasm; pore complex Molecular Function:BH domain binding; BH3 domain binding; channel activity; chaperone binding; heat shock protein binding; identical protein binding; lipid binding; protein binding; protein complex binding; protein heterodimerization activity; protein homodimerization activity Biological Process: aging; apoptosis; apoptotic mitochondrial changes; B cell apoptosis; B cell homeostasis; B cell homeostatic proliferation; B cell negative selection; blood vessel remodeling; brain development; caspase activation; caspase activation via cytochrome c; cell proliferation; cellular respiration; cerebral cortex development; development of secondary sexual characteristics; DNA damage response, signal transduction resulting in induction of apoptosis; establishment and/or maintenance of transmembrane electrochemical gradient; fertilization; germ cell development; germ cell programmed cell death; glycosphingolipid metabolic process; homeostasis of number of cells; homeostasis of number of cells within a tissue; hypothalamus development; induction of apoptosis via death domain receptors; inner mitochondrial membrane organization and biogenesis; kidney development; leukocyte homeostasis; limb morphogenesis; male gonad development; mitochondrial fragmentation during apoptosis; mitochondrial fusion; myeloid cell homeostasis; negative regulation of cell proliferation; negative regulation of fibroblast proliferation; negative regulation of neuron apoptosis; negative regulation of peptidyl-serine phosphorylation; negative regulation of protein binding; nervous system development; neuron apoptosis; neuron migration; odontogenesis of dentine-containing teeth; outer mitochondrial membrane organization and biogenesis; ovarian follicle development; positive regulation of apoptosis; positive regulation of apoptosis involved in mammary gland involution; positive regulation of B cell apoptosis; positive regulation of neuron apoptosis; positive regulation of pigmentation; positive regulation of protein oligomerization; positive regulation of release of sequestered calcium ion into cytosol; post-embryonic camera-type eye morphogenesis; post-embryonic development; protein homooligomerization; protein insertion into mitochondrial membrane during induction of apoptosis; protein oligomerization; reduction of endoplasmic reticulum calcium ion concentration; regulation of caspase activity; regulation of cell cycle; regulation of mammary gland epithelial cell proliferation; regulation of mitochondrial membrane potential; regulation of neuron apoptosis; regulation of nitrogen utilization; regulation of protein heterodimerization activity; regulation of protein homodimerization activity; release of cytochrome c from mitochondria; release of matrix enzymes from mitochondria; response to axon injury; response to cocaine; response to copper ion; response to corticosterone stimulus; response to DNA damage stimulus; response to drug; response to gamma radiation; response to ionizing radiation; response to salt stress; response to toxin; response to wounding; retina development in camera-type eye; retinal cell programmed cell death; Sertoli cell proliferation; sex differentiation; spermatid differentiation; spermatogenesis; T cell homeostatic proliferation; transformed cell apoptosis; vagina development |
| NCBI Summary: | Bcl2-related gene; involved in the regulation of apoptotic cell death [RGD, Feb 2006] |
| UniProt Code: | Q63690 |
| NCBI GenInfo Identifier: | 2493273 |
| NCBI Gene ID: | 24887 |
| NCBI Accession: | Q63690. 2 |
| UniProt Secondary Accession: | Q63690,Q62995, Q64383, |
| UniProt Related Accession: | Q63690 |
| Molecular Weight: | Da |
| NCBI Full Name: | Apoptosis regulator BAX |
| NCBI Synonym Full Names: | Bcl2-associated X protein |
| NCBI Official Symbol: | Bax |
| NCBI Protein Information: | apoptosis regulator BAX |
| UniProt Protein Name: | Apoptosis regulator BAX |
| Protein Family: | Apoptosis regulator |
| UniProt Gene Name: | Bax |
| UniProt Entry Name: | BAX_RAT |
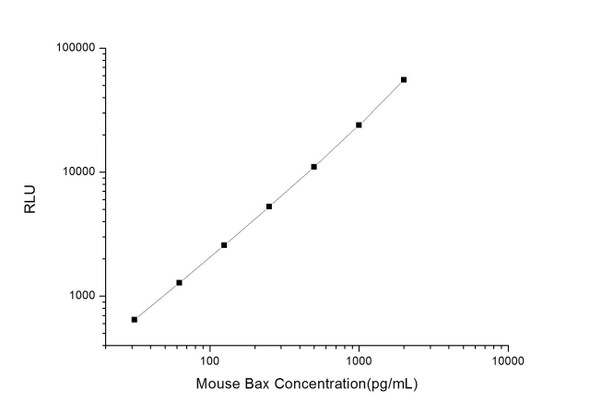
As the RLU values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (pg/mL) | RLU | Average | Corrected |
| 2000 | 51619 59823 | 55721 | 55693 |
| 1000 | 23307 24641 | 23974 | 23946 |
| 500 | 11079 10985 | 11032 | 11004 |
| 250 | 5056 5532 | 5294 | 5266 |
| 125 | 2780 2436 | 2608 | 2580 |
| 62.5 | 1315 1307 | 1311 | 1283 |
| 31.25 | 658 690 | 674 | 646 |
| 0 | 27 29 | 28 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Rat BAX were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Rat BAX were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (pg/mL) | 102.25 | 262.08 | 808.60 | 96.93 | 236.99 | 888.93 |
| Standard deviation | 9.88 | 27.49 | 88.62 | 8.83 | 17.49 | 83.91 |
| C V (%) | 9.66 | 10.49 | 10.96 | 9.11 | 7.38 | 9.44 |
Recovery
The recovery of Rat BAX spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 94-106 | 100 |
| EDTA plasma (n=5) | 89-101 | 94 |
| Cell culture media (n=5) | 92-108 | 100 |
Linearity
Samples were spiked with high concentrations of Rat BAX and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 101-117 | 103-117 | 90-105 |
| Average (%) | 108 | 110 | 97 | |
| 1:4 | Range (%) | 91-105 | 93-104 | 101-118 |
| Average (%) | 98 | 98 | 109 | |
| 1:8 | Range (%) | 92-109 | 97-108 | 85-98 |
| Average (%) | 100 | 103 | 90 | |
| 1:16 | Range (%) | 92-108 | 93-108 | 89-100 |
| Average (%) | 99 | 100 | 95 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro CLIA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent A | 1 vial, 5 mL | 4°C (shading light) |
| Substrate Reagent B | 1 vial, 5 mL | 4°C (shading light) |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100µl of standard solutions into the standard wells.
- Add 100µl of Sample / Standard dilution buffer into the control (zero) well.
- Add 100µl of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids. ) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37°C.
- Aspirate the liquid from each well, do not wash. Immediately add 100µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37°C.
- Aspirate or decant the solution from the plate and add 350µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37°C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 100µL of Substrate mixture solution to each well. Cover with a new plate seal andincubate for no more than 5 min at 37°C. Protect the plate from light.
- Determine the RLU value of each well immediately.