Mouse Cell Death ELISA Kits
Mouse TP53 (Tumor Protein 53) CLIA Kit (MOES00585)
- SKU:
- MOES00585
- Product Type:
- ELISA Kit
- ELISA Type:
- CLIA Kit
- Size:
- 96 Assays
- Sensitivity:
- 9.38pg/mL
- Range:
- 15.63-1000pg/mL
- ELISA Type:
- Sandwich
- Reactivity:
- Mouse
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Cell Death
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Mouse |
| Detection method: | Chemiluminescence |
| Detection range: | 15.63-1000 pg/mL |
| Sensitivity: | 9.38 pg/mL |
| Sample volume: | 100µL |
| Sample type: | Serum, plasma and other biological fluids |
| Repeatability: | CV < 15% |
| Specificity: | This kit recognizes Mouse TP53 in samples. No significant cross-reactivity or interference between Mouse TP53 and analogues was observed. |
This kit uses Sandwich-CLIA as the method. The micro CLIA plate provided in this kit has been pre-coated with an antibody specific to Mouse TP53. Standards or samples are added to the appropriate micro CLIA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Mouse TP53 and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Mouse TP53, biotinylated detection antibody and Avidin-HRP conjugate will appear fluorescence. The Relative light unit (RLU) value is measured spectrophotometrically by the Chemiluminescence immunoassay analyzer. The RLU value is positively associated with the concentration of Mouse TP53. The concentration of Mouse TP53 in the samples can be calculated by comparing the RLU of the samples to the standard curve.
| UniProt Protein Function: | p53: a transcription factor and major tumor suppressor that plays a major role in regulating cellular responses to DNA damage and other genomic aberrations. Activation of p53 can lead to either cell cycle arrest and DNA repair or apoptosis. More than 50 percent of human tumors contain a mutation or deletion of the TP53 gene. p53 is modified post-translationally at multiple sites. DNA damage induces phosphorylation of p53 at S15, S20 and S37, reducing its interaction with the oncoprotein MDM2. MDM2 inhibits p53 accumulation by targeting it for ubiquitination and proteasomal degradation. Phosphorylated by many kinases including Chk2 and Chk1 at S20, enhancing its tetramerization, stability and activity. The phosphorylation by CAK at S392 is increased in human tumors and has been reported to influence the growth suppressor function, DNA binding and transcriptional activation of p53. Phosphorylation of p53 at S46 regulates the ability of p53 to induce apoptosis. The acetylation of p53 appears to play a positive role in the accumulation of p53 during the stress response. Following DNA damage, p53 becomes acetylated at K382, enhancing its binding to DNA. Deacetylation of p53 can occur through interaction with SIRT1, a deacetylase that may be involved in cellular aging and the DNA damage response. p53 regulates the transcription of a set of genes encoding endosomal proteins that regulate endosomal functions. These include STEAP3 and CHMP4C, which enhance exosome production, and CAV1 and CHMP4C, which produce a more rapid endosomal clearance of the EGFR from the plasma membrane. DNA damage regulates a p53-mediated secretory pathway, increasing the secretion of some proteins such as Hsp90, SERPINE1, SERPINB5, NKEF-A, and CyPA, and inhibiting the secretion of others including CTSL and IGFBP-2. Two alternatively spliced human isoforms have been reported. Isoform 2 is expressed in quiescent lymphocytes. Seems to be non-functional. May be produced at very low levels due to a premature stop codon in the mRNA, leading to nonsense-mediated mRNA decay. |
| UniProt Protein Details: | Protein type:Tumor suppressor; Transcription factor; DNA-binding; Activator; Motility/polarity/chemotaxis; Nuclear receptor co-regulator Cellular Component: PML body; transcription factor TFIID complex; nuclear matrix; protein complex; mitochondrion; endoplasmic reticulum; replication fork; cytosol; nucleoplasm; nuclear body; transcription factor complex; mitochondrial matrix; nuclear chromatin; cytoplasm; nucleolus; intracellular; chromatin; nucleus Molecular Function:identical protein binding; protease binding; protein phosphatase 2A binding; metal ion binding; protein phosphatase binding; transcription factor binding; histone acetyltransferase binding; enzyme binding; sequence-specific DNA binding; double-stranded DNA binding; transcription factor activity; ATP binding; protein C-terminus binding; p53 binding; protein N-terminus binding; receptor tyrosine kinase binding; protein kinase binding; protein binding; histone deacetylase regulator activity; copper ion binding; DNA binding; protein heterodimerization activity; ubiquitin protein ligase binding; chaperone binding; damaged DNA binding; chromatin binding Biological Process: central nervous system development; positive regulation of apoptosis; regulation of cell cycle; positive regulation of leukocyte migration; positive regulation of transcription, DNA-dependent; multicellular organismal development; T cell differentiation in the thymus; programmed cell death; gastrulation; determination of adult life span; DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest; regulation of apoptosis; cellular response to glucose starvation; protein localization; negative regulation of neuroblast proliferation; transforming growth factor beta receptor signaling pathway; regulation of neuron apoptosis; cerebellum development; protein complex assembly; negative regulation of mitotic cell cycle; cell cycle arrest; ER overload response; response to X-ray; response to UV; response to drug; release of cytochrome c from mitochondria; somitogenesis; transcription, DNA-dependent; chromatin assembly; positive regulation of cell cycle; cell aging; circadian behavior; rRNA transcription; regulation of transcription from RNA polymerase II promoter; positive regulation of peptidyl-tyrosine phosphorylation; negative regulation of DNA replication; negative regulation of fibroblast proliferation; regulation of intracellular pH; embryonic organ development; positive regulation of transcription from RNA polymerase II promoter; response to oxidative stress; negative regulation of transcription, DNA-dependent; regulation of tissue remodeling; negative regulation of apoptosis; transcription from RNA polymerase II promoter; G1 DNA damage checkpoint; DNA damage response, signal transduction by p53 class mediator; negative regulation of smooth muscle cell proliferation; apoptosis; negative regulation of transcription from RNA polymerase II promoter; chromosome organization and biogenesis; response to salt stress; entrainment of circadian clock by photoperiod; embryonic development ending in birth or egg hatching; positive regulation of protein oligomerization; negative regulation of cell proliferation; DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator; positive regulation of histone deacetylation; regulation of transcription, DNA-dependent; T cell proliferation during immune response; regulation of catalytic activity; double-strand break repair; positive regulation of neuron apoptosis; response to gamma radiation; DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis; protein tetramerization; mitochondrial DNA repair; negative regulation of proteolysis; in utero embryonic development; B cell lineage commitment; multicellular organism growth; cell cycle; regulation of cell proliferation; T cell lineage commitment; neuron apoptosis; nucleotide-excision repair; protein import into nucleus, translocation; DNA strand renaturation; negative regulation of cell growth; negative regulation of transforming growth factor beta receptor signaling pathway; response to DNA damage stimulus |
| NCBI Summary: | This gene encodes tumor protein p53, which responds to diverse cellular stresses to regulate target genes that induce cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. p53 protein is expressed at low level in normal cells and at a high level in a variety of transformed cell lines, where it's believed to contribute to transformation and malignancy. p53 is a DNA-binding protein containing transcription activation, DNA-binding, and oligomerization domains. It is postulated to bind to a p53-binding site and activate expression of downstream genes that inhibit growth and/or invasion, and thus function as a tumor suppressor. Mice deficient for this gene are developmentally normal but are susceptible to spontaneous tumors. Evidence to date shows that this gene contains one promoter, in contrast to alternative promoters of the human gene, and transcribes a few of splice variants which encode different isoforms, although the biological validity or the full-length nature of some variants has not been determined. [provided by RefSeq, Jul 2008] |
| UniProt Code: | P02340 |
| NCBI GenInfo Identifier: | 172047304 |
| NCBI Gene ID: | 22059 |
| NCBI Accession: | P02340. 3 |
| UniProt Secondary Accession: | P02340,Q9QUP3, |
| UniProt Related Accession: | P02340 |
| Molecular Weight: | 393 |
| NCBI Full Name: | Cellular tumor antigen p53 |
| NCBI Synonym Full Names: | transformation related protein 53 |
| NCBI Official Symbol: | Trp53 |
| NCBI Official Synonym Symbols: | bbl; bfy; bhy; p44; p53; Tp53 |
| NCBI Protein Information: | cellular tumor antigen p53; tumor supressor p53; tumor suppressor p53; p53 cellular tumor antigen |
| UniProt Protein Name: | Cellular tumor antigen p53 |
| UniProt Synonym Protein Names: | Tumor suppressor p53 |
| Protein Family: | TP53-regulating kinase |
| UniProt Gene Name: | Tp53 |
| UniProt Entry Name: | P53_MOUSE |
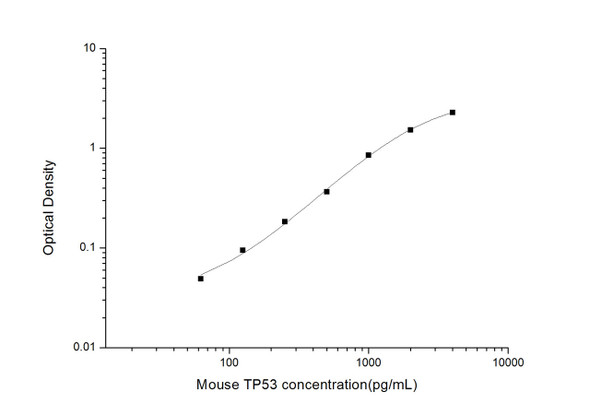
As the RLU values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (pg/mL) | RLU | Average | Corrected |
| 1000 | 51432 55918 | 53675 | 53650 |
| 500 | 20249 24729 | 22489 | 22464 |
| 250 | 10823 9599 | 10211 | 10186 |
| 125 | 4541 5261 | 4901 | 4876 |
| 62.5 | 2618 2288 | 2453 | 2428 |
| 31.25 | 1366 1196 | 1281 | 1256 |
| 15.63 | 669 747 | 708 | 683 |
| 0 | 24 26 | 25 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Mouse TP53 were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Mouse TP53 were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (pg/mL) | 54.74 | 91.82 | 364.25 | 57.10 | 99.30 | 400.50 |
| Standard deviation | 6.36 | 7.42 | 39.81 | 4.65 | 10.23 | 30.80 |
| C V (%) | 11.62 | 8.08 | 10.93 | 8.14 | 10.30 | 7.69 |
Recovery
The recovery of Mouse TP53 spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 92-106 | 98 |
| EDTA plasma (n=5) | 86-101 | 93 |
| Cell culture media (n=5) | 89-102 | 96 |
Linearity
Samples were spiked with high concentrations of Mouse TP53 and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 94-107 | 91-103 | 91-106 |
| Average (%) | 100 | 97 | 99 | |
| 1:4 | Range (%) | 101-117 | 92-103 | 99-115 |
| Average (%) | 109 | 97 | 105 | |
| 1:8 | Range (%) | 85-99 | 97-112 | 104-116 |
| Average (%) | 91 | 103 | 110 | |
| 1:16 | Range (%) | 100-118 | 97-113 | 92-107 |
| Average (%) | 108 | 105 | 99 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro CLIA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent A | 1 vial, 5 mL | 4°C (shading light) |
| Substrate Reagent B | 1 vial, 5 mL | 4°C (shading light) |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100 µL of standard solutions into the standard wells.
- Add 100 µL of Sample / Standard dilution buffer into the control (zero) well.
- Add 100 µL of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids. ) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37 °C.
- Aspirate the liquid from each well, do not wash. Immediately add 100 µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37 °C.
- Aspirate or decant the solution from the plate and add 350 µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100 µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37 °C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 100 µL of Substrate mixture solution to each well. Cover with a new plate seal andincubate for no more than 5 min at 37 °C. Protect the plate from light.
- Determine the RLU value of each well immediately.