Mouse Cell Death ELISA Kits
Mouse SIRT1 (NAD-dependent protein deacetylase sirtuin-1) ELISA Kit (MOES00911)
- SKU:
- MOES00911
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- Q923E4
- Sensitivity:
- 0.09ng/mL
- Range:
- 0.16-10ng/mL
- ELISA Type:
- Sandwich
- Synonyms:
- SIR2L1,SIR2alpha,sir2-like 1,sirtuin 1,Silent Mating Type Information Regulation 2 Homolog 1
- Reactivity:
- Mouse
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Cell Death
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Mouse |
| Detection Method: | Colormetric |
| Detection Range: | 0.16-10 ng/mL |
| Sensitivity: | 0.10 ng/mL |
| Sample Volume Required Per Well: | 100µL |
| Sample Type: | Serum, plasma and other biological fluids |
| Specificity: | This kit recognizes Mouse SIRT1 in samples. No significant cross-reactivity or interference between Mouse SIRT1 and analogues was observed. |
This ELISA kit uses Sandwich-ELISA as the method. The micro ELISA plate provided in this kit has been pre-coated with an antibody specific to Mouse SIRT1. Standards or samples are added to the appropriate micro ELISA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Mouse SIRT1 and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Mouse SIRT1, biotinylated detection antibody and Avidin-HRP conjugate will appear blue in color. The enzyme-substrate reaction is terminated by adding Stop Solution and the color turns yellow. The optical density (OD) is measured spectrophotometrically at a wavelength of 450 nm ± 2 nm. The OD value is proportional to the concentration of Mouse SIRT1. The concentration of Mouse SIRT1 in samples can be calculated by comparing the OD of the samples to the standard curve.
| UniProt Protein Function: | SIRT1: an NAD-dependent protein deacetylase that links transcriptional regulation directly to intracellular energetics and participates in the coordination of several separate cellular functions such as cell cycle, response to DNA damage, metobolism, apoptosis and autophagy. Deacetylates a broad range of transcription factors and coregulators, thereby regulating target gene expression positively and negatively. Serves as a sensor of the cytosolic ratio of NAD(+)/NADH which is altered by glucose deprivation and metabolic changes associated with caloric restriction. Essential in skeletal muscle cell differentiation and in response to low nutrients mediates the inhibitory effect on skeletal myoblast differentiation which also involves 5'-AMP-activated protein kinase (AMPK) and nicotinamide phosphoribosyltransferase (NAMPT). Component of the eNoSC (energy-dependent nucleolar silencing) complex, a complex that mediates silencing of rDNA in response to intracellular energy status and acts by recruiting histone-modifying enzymes. Elevation of NAD(+)/NADP(+) ratio activates SIRT1. Recruited to LRH1 target gene promoters by NR0B2/SHP thereby stimulating histone H3 and H4 deacetylation leading to transcriptional repression. Implicated in regulation of adipogenesis and fat mobilization in white adipocytes by repression of PPARG. Involved in liver and muscle metabolism. Is involved in autophagy, presumably by deacetylating ATG5, ATG7 and ATG8. Deacetylates AKT1 which leads to enhanced binding of AKT1 and PDK1 to PIP3 and promotes their activation. Widely expressed. Inhibited by nicotinamide. Belongs to the sirtuin family. Class I subfamily. 2 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:Nuclear receptor co-regulator; Apoptosis; Deacetylase; EC 3. 5. 1. - Cellular Component: axon; chromatin; chromatin silencing complex; cytoplasm; cytosol; ESC/E(Z) complex; growth cone; mitochondrion; nuclear chromatin; nuclear envelope; nuclear heterochromatin; nuclear inner membrane; nucleoplasm; nucleus; PML body Molecular Function:bHLH transcription factor binding; deacetylase activity; enzyme binding; histone binding; histone deacetylase activity; HLH domain binding; identical protein binding; mitogen-activated protein kinase binding; NAD-dependent histone deacetylase activity; NAD-dependent histone deacetylase activity (H3-K9 specific); nuclear hormone receptor binding; p53 binding; protein binding; protein C-terminus binding; protein deacetylase activity; protein domain specific binding; protein kinase B binding; transcription corepressor activity; transcription factor binding Biological Process: angiogenesis; behavioral response to starvation; cell glucose homeostasis; cellular response to starvation; cholesterol homeostasis; chromatin silencing at rDNA; circadian regulation of gene expression; DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis; DNA damage response, signal transduction resulting in induction of apoptosis; DNA synthesis during DNA repair; establishment and/or maintenance of chromatin architecture; establishment of chromatin silencing; fatty acid homeostasis; histone deacetylation; inhibition of NF-kappaB transcription factor; leptin-mediated signaling pathway; macrophage differentiation; maintenance of chromatin silencing; negative regulation of apoptosis; negative regulation of cell growth; negative regulation of DNA binding; negative regulation of DNA damage response, signal transduction by p53 class mediator; negative regulation of fat cell differentiation; negative regulation of growth hormone secretion; negative regulation of helicase activity; negative regulation of I-kappaB kinase/NF-kappaB cascade; negative regulation of neuron apoptosis; negative regulation of phosphorylation; negative regulation of prostaglandin biosynthetic process; negative regulation of protein kinase B signaling cascade; negative regulation of TOR signaling pathway; negative regulation of transcription factor activity; negative regulation of transcription from RNA polymerase II promoter; negative regulation of transcription, DNA-dependent; negative regulation of transforming growth factor beta receptor signaling pathway; negative regulation of tumor necrosis factor production; ovulation from ovarian follicle; peptidyl-lysine acetylation; positive regulation of adaptive immune response; positive regulation of angiogenesis; positive regulation of apoptosis; positive regulation of caspase activity; positive regulation of cell proliferation; positive regulation of chromatin silencing; positive regulation of DNA repair; positive regulation of endothelial cell proliferation; positive regulation of gluconeogenesis; positive regulation of heart rate; positive regulation of histone H3-K9 methylation; positive regulation of insulin receptor signaling pathway; positive regulation of macroautophagy; positive regulation of MHC class II biosynthetic process; positive regulation of phosphoinositide 3-kinase cascade; positive regulation of protein amino acid phosphorylation; positive regulation of skeletal muscle cell proliferation; positive regulation of transcription from RNA polymerase II promoter; positive regulation of vasodilation; proteasomal ubiquitin-dependent protein catabolic process; protein amino acid deacetylation; protein destabilization; protein ubiquitination; pyrimidine dimer repair via nucleotide-excision repair; regulation of cell proliferation; regulation of endodeoxyribonuclease activity; regulation of mitotic cell cycle; regulation of protein import into nucleus, translocation; response to DNA damage stimulus; response to ethanol; response to hydrogen peroxide; response to insulin stimulus; response to oxidative stress; single strand break repair; spermatogenesis; triacylglycerol mobilization; white fat cell differentiation |
| NCBI Summary: | This gene encodes a member of the sirtuin family of proteins, characterized by their deacetylase activity and proposed role in longevity. The encoded protein regulates gene expression in a wide range of cell and tissue types through its NAD+-dependent deacetylation of histones, transcription factors and transcriptional coactivators. Brain-specific overexpression of this gene has been shown to result in increased median lifespan. Viability of homozygous knockout mice for this gene varies with strain background. Homozygous knockout mice of strains that do not exhibit embryonic lethality are sterile and have a reduced lifespan. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2015] |
| UniProt Code: | Q923E4 |
| NCBI GenInfo Identifier: | 38258625 |
| NCBI Gene ID: | 93759 |
| NCBI Accession: | Q923E4. 2 |
| UniProt Secondary Accession: | Q923E4,Q9QXG8, |
| UniProt Related Accession: | Q923E4 |
| Molecular Weight: | 59,875 Da |
| NCBI Full Name: | NAD-dependent protein deacetylase sirtuin-1 |
| NCBI Synonym Full Names: | sirtuin 1 |
| NCBI Official Symbol: | Sirt1 |
| NCBI Official Synonym Symbols: | Sir2; Sir2a; SIR2L1; AA673258; Sir2alpha |
| NCBI Protein Information: | NAD-dependent protein deacetylase sirtuin-1 |
| UniProt Protein Name: | NAD-dependent protein deacetylase sirtuin-1 |
| UniProt Synonym Protein Names: | Regulatory protein SIR2 homolog 1; SIR2-like protein 1; SIR2alpha; Sir2; mSIR2a |
| Protein Family: | NAD-dependent protein deacetylase sirtuin |
| UniProt Gene Name: | Sirt1 |
| UniProt Entry Name: | SIR1_MOUSE |
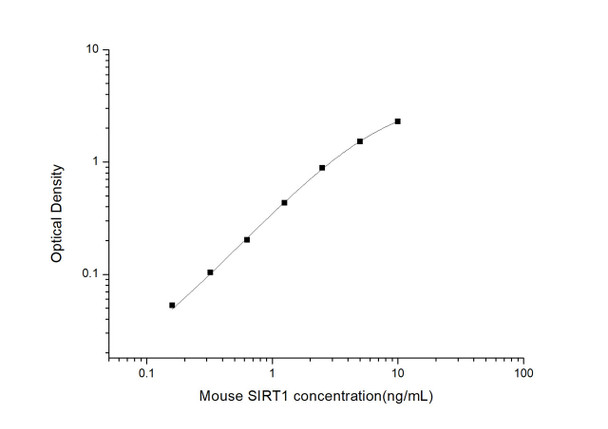
As the OD values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (ng/mL) | O.D | Average | Corrected |
| 10 | 2.364 2.386 | 2.375 | 2.303 |
| 5 | 1.563 1.623 | 1.593 | 1.521 |
| 2.5 | 0.965 0.947 | 0.956 | 0.884 |
| 1.25 | 0.485 0.523 | 0.504 | 0.432 |
| 0.63 | 0.276 0.272 | 0.274 | 0.202 |
| 0.32 | 0.19 0.162 | 0.176 | 0.104 |
| 0.16 | 0.117 0.133 | 0.125 | 0.053 |
| 0 | 0.066 0.078 | 0.072 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Mouse SIRT1 were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Mouse SIRT1 were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (ng/mL) | 0.48 | 0.90 | 4.84 | 0.46 | 0.81 | 4.36 |
| Standard deviation | 0.02 | 0.05 | 0.22 | 0.03 | 0.05 | 0.23 |
| C V (%) | 4.17 | 5.56 | 4.55 | 6.52 | 6.17 | 5.28 |
Recovery
The recovery of Mouse SIRT1 spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 94-105 | 99 |
| EDTA plasma (n=5) | 83-96 | 90 |
| Cell culture media (n=5) | 94-109 | 100 |
Linearity
Samples were spiked with high concentrations of Mouse SIRT1 and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 87-101 | 89-102 | 85-97 |
| Average (%) | 93 | 95 | 92 | |
| 1:4 | Range (%) | 90-106 | 84-96 | 85-97 |
| Average (%) | 97 | 90 | 92 | |
| 1:8 | Range (%) | 88-101 | 84-94 | 88-102 |
| Average (%) | 95 | 89 | 94 | |
| 1:16 | Range (%) | 89-105 | 81-95 | 85-99 |
| Average (%) | 96 | 87 | 90 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro ELISA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent | 1 vial, 10 mL | 4°C(shading light) |
| Stop Solution | 1 vial, 10 mL | 4°C |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100µl of standard solutions into the standard wells.
- Add 100µl of Sample / Standard dilution buffer into the control (zero) well.
- Add 100µl of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37°C.
- Aspirate the liquid from each well, do not wash. Immediately add 100µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37°C.
- Aspirate or decant the solution from the plate and add 350µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37°C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 90µL of Substrate Reagent to each well. Cover with a new plate seal and incubate forapproximately 15 min at 37°C. Protect the plate from light. Note: the reaction time can beshortened or extended according to the actual color change, but not by more than 30min.
- Add 50 µL of Stop Solution to each well. Note: Adding the stop solution should be done inthe same order as the substrate solution.
- Determine the optical density (OD value) of each well immediately with a microplate readerset at 450 nm.