Description
| Product Name: | Lamin A/C (Phospho-Ser22) Colorimetric Cell-Based ELISA |
| Product Code: | CBCAB00504 |
| ELISA Type: | Cell-Based |
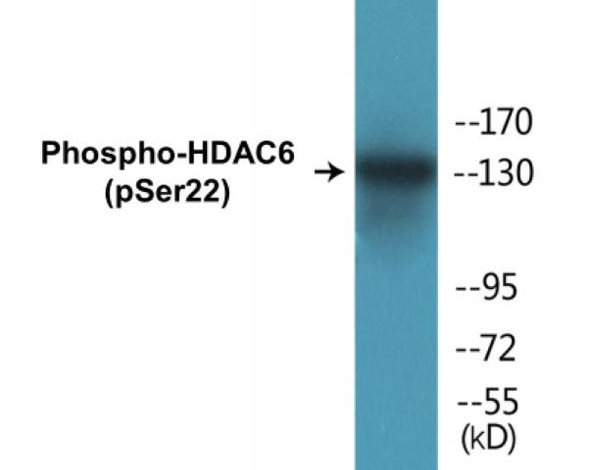
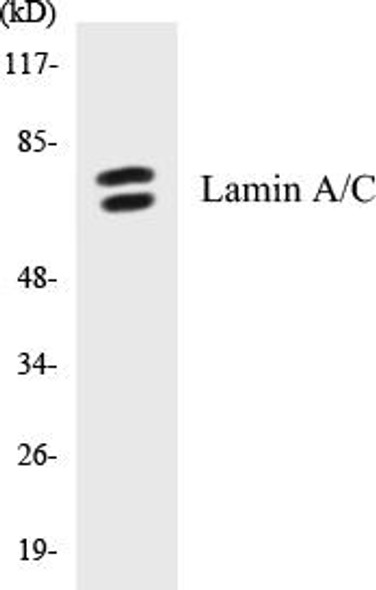
| Target: | Lamin A/C (Phospho-Ser22) |
| Reactivity: | Human, Mouse, Rat |
| Dynamic Range: | > 5000 Cells |
| Detection Method: | Colorimetric 450 nm |
| Format: | 2 x 96-Well Microplates |
The Lamin A/C (Phospho-Ser22) Colorimetric Cell-Based ELISA Kit is a convenient, lysate-free, high throughput and sensitive assay kit that can detect Lamin A/C protein phosphorylation and expression profile in cells. The kit can be used for measuring the relative amounts of phosphorylated Lamin A/C in cultured cells as well as screening for the effects that various treatments, inhibitors (ie. siRNA or chemicals), or activators have on Lamin A/C phosphorylation.
Qualitative determination of Lamin A/C (Phospho-Ser22) concentration is achieved by an indirect ELISA format. In essence, Lamin A/C (Phospho-Ser22) is captured by Lamin A/C (Phospho-Ser22)-specific primary antibodies while the HRP-conjugated secondary antibodies bind the Fc region of the primary antibody. Through this binding, the HRP enzyme conjugated to the secondary antibody can catalyze a colorimetric reaction upon substrate addition. Due to the qualitative nature of the Cell-Based ELISA, multiple normalization methods are needed:
| 1. | A monoclonal antibody specific for human GAPDH is included to serve as an internal positive control in normalizing the target absorbance values. |
| 2. | Following the colorimetric measurement of HRP activity via substrate addition, the Crystal Violet whole-cell staining method may be used to determine cell density. After staining, the results can be analysed by normalizing the absorbance values to cell amounts, by which the plating difference can be adjusted. |
| Database Information: | Gene ID: 4000, UniProt ID: P02545, OMIM: 115200/150330/151660/159001/176670/181350/248370/275210/277700/604929/605588/608056/611618, Unigene: Hs.594444 |
| Gene Symbol: | LMNA |
| Sub Type: | Phospho |
| UniProt Protein Function: | Function: Lamins are components of the nuclear lamina, a fibrous layer on the nucleoplasmic side of the inner nuclear membrane, which is thought to provide a framework for the nuclear envelope and may also interact with chromatin. Lamin A and C are present in equal amounts in the lamina of mammals. Plays an important role in nuclear assembly, chromatin organization, nuclear membrane and telomere dynamics. Ref.24 Ref.25Prelamin-A/C can accelerate smooth muscle cell senescence. It acts to disrupt mitosis and induce DNA damage in vascular smooth muscle cells (VSMCs), leading to mitotic failure, genomic instability, and premature senescence. Ref.24 Ref.25 |
| UniProt Protein Details: | Subunit structure: Homodimer of lamin A and lamin C. Interacts with lamin-associated polypeptides IA, IB and TMPO-alpha, RB1 and with emerin. Interacts with SREBF1, SREBF2, SUN2 and TMEM43 By similarity. Proteolytically processed isoform A interacts with NARF. Interacts with SUN1. Prelamin-A/C interacts with EMD. Interacts with MLIP; may regulate MLIP localization to the nucleus envelope. Interacts with DMPK; may regulate nuclear envelope stability. Ref.12 Ref.13 Ref.22 Ref.26 Ref.29 Ref.30 Subcellular location: Nucleus. Nucleus envelope. Note: Farnesylation of prelamin-A/C facilitates nuclear envelope targeting and subsequent cleaveage by ZMPSTE24/FACE1 to remove the farnesyl group produces mature lamin-A/C, which can then be inserted into the nuclear lamina. EMD is required for proper localization of non-farnesylated prelamin-A/C. Ref.17 Ref.22 Tissue specificity: In the arteries, prelamin-A/C accumulation is not observed in young healthy vessels but is prevalent in medial vascular smooth muscle cells (VSMCs) from aged individuals and in atherosclerotic lesions, where it often colocalizes with senescent and degenerate VSMCs. Prelamin-A/C expression increases with age and disease. In normal aging, the accumulation of prelamin-A/C is caused in part by the down-regulation of ZMPSTE24/FACE1 in response to oxidative stress. Ref.25 Post-translational modification: Increased phosphorylation of the lamins occurs before envelope disintegration and probably plays a role in regulating lamin associations.Proteolytic cleavage of the C-terminal of 18 residues of prelamin-A/C results in the production of lamin-A/C. The prelamin-A/C maturation pathway includes farnesylation of CAAX motif, ZMPSTE24/FACE1 mediated cleavage of the last three amino acids, methylation of the C-terminal cysteine and endoproteolytic removal of the last 15 C-terminal amino acids. Proteolytic cleavage requires prior farnesylation and methylation, and absence of these blocks cleavage.Sumoylation is necessary for the localization to the nuclear envelope. Ref.17Farnesylation of prelamin-A/C facilitates nuclear envelope targeting. Involvement in Disease: Emery-Dreifuss muscular dystrophy 2, autosomal dominant (EDMD2) [MIM:181350]: A form of Emery-Dreifuss muscular dystrophy, a degenerative myopathy characterized by weakness and atrophy of muscle without involvement of the nervous system, early contractures of the elbows, Achilles tendons and spine, and cardiomyopathy associated with cardiac conduction defects.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.26 Ref.35 Ref.39 Ref.40 Ref.44 Ref.45 Ref.47 Ref.55 Ref.64 Ref.67 Ref.73 Ref.77 Ref.87Emery-Dreifuss muscular dystrophy 3, autosomal recessive (EDMD3) [MIM:181350]: A form of Emery-Dreifuss muscular dystrophy, a degenerative myopathy characterized by weakness and atrophy of muscle without involvement of the nervous system, early contractures of the elbows, Achilles tendons and spine, and cardiomyopathy associated with cardiac conduction defects.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.88Cardiomyopathy, dilated 1A (CMD1A) [MIM:115200]: A disorder characterized by ventricular dilation and impaired systolic function, resulting in congestive heart failure and arrhythmia. Patients are at risk of premature death.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.17 Ref.36 Ref.46 Ref.47 Ref.49 Ref.54 Ref.59 Ref.63 Ref.64 Ref.68 Ref.69 Ref.78 Ref.86Lipodystrophy, familial partial, 2 (FPLD2) [MIM:151660]: A disorder characterized by the loss of subcutaneous adipose tissue in the lower parts of the body (limbs, buttocks, trunk). It is accompanied by an accumulation of adipose tissue in the face and neck causing a double chin, fat neck, or cushingoid appearance. Adipose tissue may also accumulate in the axillae, back, labia majora, and intraabdominal region. Affected patients are insulin-resistant and may develop glucose intolerance and diabetes mellitus after age 20 years, hypertriglyceridemia, and low levels of high density lipoprotein cholesterol.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.37 Ref.41 Ref.43 Ref.47 Ref.53 Ref.56 Ref.61 Ref.81Limb-girdle muscular dystrophy 1B (LGMD1B) [MIM:159001]: An autosomal dominant degenerative myopathy with age-related atrioventricular cardiac conduction disturbances, dilated cardiomyopathy, and the absence of early contractures. Characterized by slowly progressive skeletal muscle weakness of the hip and shoulder girdles. Muscle biopsy shows mild dystrophic changes.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.42 Ref.48 Ref.55 Ref.58 Ref.77 Ref.83Charcot-Marie-Tooth disease 2B1 (CMT2B1) [MIM:605588]: A recessive axonal form of Charcot-Marie-Tooth disease, a disorder of the peripheral nervous system, characterized by progressive weakness and atrophy, initially of the peroneal muscles and later of the distal muscles of the arms. Charcot-Marie-Tooth disease is classified in two main groups on the basis of electrophysiologic properties and histopathology: primary peripheral demyelinating neuropathies (designated CMT1 when they are dominantly inherited) and primary peripheral axonal neuropathies (CMT2). Neuropathies of the CMT2 group are characterized by signs of axonal regeneration in the absence of obvious myelin alterations, normal or slightly reduced nerve conduction velocities, and progressive distal muscle weakness and atrophy. Nerve conduction velocities are normal or slightly reduced.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.50Hutchinson-Gilford progeria syndrome (HGPS) [MIM:176670]: Rare genetic disorder characterized by features reminiscent of marked premature aging.Note: The disease is caused by mutations affecting the gene represented in this entry. HGPS is caused by the toxic accumulation of a mutant form of lamin-A/C. This mutant protein, called progerin, acts to deregulate mitosis and DNA damage signaling, leading to premature cell death and senescence. Progerin lacks the conserved ZMPSTE24/FACE1 cleavage site and therefore remains permanently farnesylated. Thus, although it can enter the nucleus and associate with the nuclear envelope, it cannot incorporate normally into the nuclear lamina. Ref.26 Ref.62 Ref.65 Ref.66 Ref.72 Ref.74Cardiomyopathy, dilated, with hypergonadotropic hypogonadism (CMDHH) [MIM:212112]: A disorder characterized by the association of genital anomalies, hypergonadotropic hypogonadism and dilated cardiomyopathy. Patients can present other variable clinical manifestations including mental retardation, skeletal anomalies, scleroderma-like skin, graying and thinning of hair, osteoporosis. Dilated cardiomyopathy is characterized by ventricular dilation and impaired systolic function, resulting in congestive heart failure and arrhythmia.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.65 Ref.80 Ref.85Mandibuloacral dysplasia with type A lipodystrophy (MADA) [MIM:248370]: A disorder characterized by mandibular and clavicular hypoplasia, acroosteolysis, delayed closure of the cranial suture, progeroid appearance, partial alopecia, soft tissue calcinosis, joint contractures, and partial lipodystrophy with loss of subcutaneous fat from the extremities. Adipose tissue in the face, neck and trunk is normal or increased.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.52 Ref.76 Ref.79Lethal tight skin contracture syndrome (LTSCS) [MIM:275210]: Rare disorder mainly characterized by intrauterine growth retardation, tight and rigid skin with erosions, prominent superficial vasculature and epidermal hyperkeratosis, facial features (small mouth, small pinched nose and micrognathia), sparse/absent eyelashes and eyebrows, mineralization defects of the skull, thin dysplastic clavicles, pulmonary hypoplasia, multiple joint contractures and an early neonatal lethal course. Liveborn children usually die within the first week of life. The overall prevalence of consanguineous cases suggested an autosomal recessive inheritance.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.70Heart-hand syndrome Slovenian type (HHS-Slovenian) [MIM:610140]: Heart-hand syndrome (HHS) is a clinically and genetically heterogeneous disorder characterized by the co-occurrence of a congenital cardiac disease and limb malformations.Note: The disease is caused by mutations affecting the gene represented in this entry.Muscular dystrophy congenital LMNA-related (MDCL) [MIM:613205]: A form of congenital muscular dystrophy. Patients present at birth, or within the first few months of life, with hypotonia, muscle weakness and often with joint contractures.Note: The disease is caused by mutations affecting the gene represented in this entry. Ref.84 Miscellaneous: There are three types of lamins in human cells: A, B, and C.The structural integrity of the lamina is strictly controlled by the cell cycle, as seen by the disintegration and formation of the nuclear envelope in prophase and telophase, respectively. Sequence similarities: Belongs to the intermediate filament family. Sequence caution: The sequence CAA27173.1 differs from that shown. Reason: Frameshift at position 582. |
| NCBI Summary: | The nuclear lamina consists of a two-dimensional matrix of proteins located next to the inner nuclear membrane. The lamin family of proteins make up the matrix and are highly conserved in evolution. During mitosis, the lamina matrix is reversibly disassembled as the lamin proteins are phosphorylated. Lamin proteins are thought to be involved in nuclear stability, chromatin structure and gene expression. Vertebrate lamins consist of two types, A and B. Alternative splicing results in multiple transcript variants. Mutations in this gene lead to several diseases: Emery-Dreifuss muscular dystrophy, familial partial lipodystrophy, limb girdle muscular dystrophy, dilated cardiomyopathy, Charcot-Marie-Tooth disease, and Hutchinson-Gilford progeria syndrome. [provided by RefSeq, Apr 2012] |
| UniProt Code: | P02545 |
| NCBI GenInfo Identifier: | 125962 |
| NCBI Gene ID: | 4000 |
| NCBI Accession: | P02545.1 |
| UniProt Secondary Accession: | P02545,P02546, Q5TCJ2, Q5TCJ3, Q969I8, Q96JA2, B4DI32 D3DVB0, E7EUI9, |
| UniProt Related Accession: | P02545 |
| Molecular Weight: | 74,139 Da |
| NCBI Full Name: | Prelamin-A/C |
| NCBI Synonym Full Names: | lamin A/C |
| NCBI Official Symbol: | LMNA |
| NCBI Official Synonym Symbols: | FPL; IDC; LFP; CDDC; EMD2; FPLD; HGPS; LDP1; LMN1; LMNC; PRO1; CDCD1; CMD1A; FPLD2; LMNL1; CMT2B1; LGMD1B |
| NCBI Protein Information: | lamin; 70 kDa lamin; prelamin-A/C; lamin A/C-like 1; renal carcinoma antigen NY-REN-32 |
| UniProt Protein Name: | Prelamin-A/C |
| UniProt Synonym Protein Names: | 70 kDa lamin; Renal carcinoma antigen NY-REN-32 |
| Protein Family: | Prelamin |
| UniProt Gene Name: | LMNA |
| UniProt Entry Name: | LMNA_HUMAN |
| Component | Quantity |
| 96-Well Cell Culture Clear-Bottom Microplate | 2 plates |
| 10X TBS | 24 mL |
| Quenching Buffer | 24 mL |
| Blocking Buffer | 50 mL |
| 15X Wash Buffer | 50 mL |
| Primary Antibody Diluent | 12 mL |
| 100x Anti-Phospho Target Antibody | 60 µL |
| 100x Anti-Target Antibody | 60 µL |
| Anti-GAPDH Antibody | 60 µL |
| HRP-Conjugated Anti-Rabbit IgG Antibody | 12 mL |
| HRP-Conjugated Anti-Mouse IgG Antibody | 12 mL |
| SDS Solution | 12 mL |
| Stop Solution | 24 mL |
| Ready-to-Use Substrate | 12 mL |
| Crystal Violet Solution | 12 mL |
| Adhesive Plate Seals | 2 seals |
The following materials and/or equipment are NOT provided in this kit but are necessary to successfully conduct the experiment:
- Microplate reader able to measure absorbance at 450 nm and/or 595 nm for Crystal Violet Cell Staining (Optional)
- Micropipettes with capability of measuring volumes ranging from 1 µL to 1 ml
- 37% formaldehyde (Sigma Cat# F-8775) or formaldehyde from other sources
- Squirt bottle, manifold dispenser, multichannel pipette reservoir or automated microplate washer
- Graph paper or computer software capable of generating or displaying logarithmic functions
- Absorbent papers or vacuum aspirator
- Test tubes or microfuge tubes capable of storing ≥1 ml
- Poly-L-Lysine (Sigma Cat# P4832 for suspension cells)
- Orbital shaker (optional)
- Deionized or sterile water
*Note: Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
| Step | Procedure |
| 1. | Seed 200 µL of 20,000 adherent cells in culture medium in each well of a 96-well plate. The plates included in the kit are sterile and treated for cell culture. For suspension cells and loosely attached cells, coat the plates with 100 µL of 10 µg/ml Poly-L-Lysine (not included) to each well of a 96-well plate for 30 minutes at 37°C prior to adding cells. |
| 2. | Incubate the cells for overnight at 37°C, 5% CO2. |
| 3. | Treat the cells as desired. |
| 4. | Remove the cell culture medium and rinse with 200 µL of 1x TBS, twice. |
| 5. | Fix the cells by incubating with 100 µL of Fixing Solution for 20 minutes at room temperature. The 4% formaldehyde is used for adherent cells and 8% formaldehyde is used for suspension cells and loosely attached cells. |
| 6. | Remove the Fixing Solution and wash the plate 3 times with 200 µL 1x Wash Buffer for five minutes each time with gentle shaking on the orbital shaker. The plate can be stored at 4°C for a week. |
| 7. | Add 100 µL of Quenching Buffer and incubate for 20 minutes at room temperature. |
| 8. | Wash the plate 3 times with 1x Wash Buffer for 5 minutes each time. |
| 9. | Add 200 µL of Blocking Buffer and incubate for 1 hour at room temperature. |
| 10. | Wash 3 times with 200 µL of 1x Wash Buffer for 5 minutes each time. |
| 11. | Add 50 µL of 1x primary antibodies Anti-Lamin A/C (Phospho-Ser22) Antibody, Anti-Lamin A/C Antibody and/or Anti-GAPDH Antibody) to the corresponding wells, cover with Parafilm and incubate for 16 hours (overnight) at 4°C. If the target expression is known to be high, incubate for 2 hours at room temperature. |
| 12. | Wash 3 times with 200 µL of 1x Wash Buffer for 5 minutes each time. |
| 13. | Add 50 µL of 1x secondary antibodies (HRP-Conjugated AntiRabbit IgG Antibody or HRP-Conjugated Anti-Mouse IgG Antibody) to corresponding wells and incubate for 1.5 hours at room temperature. |
| 14. | Wash 3 times with 200 µL of 1x Wash Buffer for 5 minutes each time. |
| 15. | Add 50 µL of Ready-to-Use Substrate to each well and incubate for 30 minutes at room temperature in the dark. |
| 16. | Add 50 µL of Stop Solution to each well and read OD at 450 nm immediately using the microplate reader. |
(Additional Crystal Violet staining may be performed if desired – details of this may be found in the kit technical manual.)