Human Epigenetics and Nuclear Signaling ELISA Kits
Human SMN1 (Survival of Motor Neuron 1, Telomeric) CLIA Kit (HUES00979)
- SKU:
- HUES00979
- Product Type:
- ELISA Kit
- ELISA Type:
- CLIA Kit
- Size:
- 96 Assays
- Sensitivity:
- 9.38pg/mL
- Range:
- 15.63-1000pg/mL
- ELISA Type:
- Sandwich
- Synonyms:
- BCD541, Gemin-1, SMA, SMNT, T-BCD541, TDRD16A
- Reactivity:
- Human
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Epigenetics and Nuclear Signaling
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Human |
| Detection method: | Chemiluminescence |
| Detection range: | 15.63-1000 pg/mL |
| Sensitivity: | 9.38 pg/mL |
| Sample volume: | 100µL |
| Sample type: | Serum, plasma and other biological fluids |
| Repeatability: | CV < 15% |
| Specificity: | This kit recognizes Human SMN1 in samples. No significant cross-reactivity or interference between Human SMN1 and analogues was observed. |
This kit uses Sandwich-CLIA as the method. The micro CLIA plate provided in this kit has been pre-coated with an antibody specific to Human SMN1. Standards or samples are added to the appropriate micro CLIA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Human SMN1 and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Human SMN1, biotinylated detection antibody and Avidin-HRP conjugate will appear fluorescence. The Relative light unit (RLU) value is measured spectrophotometrically by the Chemiluminescence immunoassay analyzer. The RLU value is positively associated with the concentration of Human SMN1. The concentration of Human SMN1 in the samples can be calculated by comparing the RLU of the samples to the standard curve.
| UniProt Protein Function: | SMN: The SMN complex plays an essential role in spliceosomal snRNP assembly in the cytoplasm and is required for pre-mRNA splicing in the nucleus. It may also play a role in the metabolism of snoRNPs. Defects in SMN1 are the cause of spinal muscular atrophy autosomal recessive type 1 (SMA1). Spinal muscular atrophy refers to a group of neuromuscular disorders characterized by degeneration of the anterior horn cells of the spinal cord, leading to symmetrical muscle weakness and atrophy. Autosomal recessive forms are classified according to the age of onset, the maximum muscular activity achieved, and survivorship. The severity of the disease is mainly determined by the copy number of SMN2, a copy gene which predominantly produces exon 7-skipped transcripts and only low amount of full-length transcripts that encode for a protein identical to SMN1. Only about 4% of SMA patients bear one SMN1 copy with an intragenic mutation. SMA1 is a severe form, with onset before 6 months of age. SMA1 patients never achieve the ability to sit. Defects in SMN1 are the cause of spinal muscular atrophy autosomal recessive type 2 (SMA2). SMA2 is an autosomal recessive spinal muscular atrophy of intermediate severity, with onset between 6 and 18 months. Patients do not reach the motor milestone of standing, and survive into adulthood. Defects in SMN1 are the cause of spinal muscular atrophy autosomal recessive type 3 (SMA3). SMA3 is an autosomal recessive spinal muscular atrophy with onset after 18 months. SMA3 patients develop ability to stand and walk and survive into adulthood. Defects in SMN1 are the cause of spinal muscular atrophy autosomal recessive type 4 (SMA4). SMA4 is an autosomal recessive spinal muscular atrophy characterized by symmetric proximal muscle weakness with onset in adulthood and slow disease progression. SMA4 patients can stand and walk. Belongs to the SMN family. 4 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:RNA processing; RNA-binding Chromosomal Location of Human Ortholog: 5q13. 2 Cellular Component: Cajal body; cytoplasm; cytosol; neuron projection; nucleoplasm; nucleus; perikaryon; SMN complex Molecular Function:identical protein binding; protein binding Biological Process: nuclear import; spliceosomal snRNP biogenesis; spliceosome assembly; transcription termination Disease: Spinal Muscular Atrophy, Type I; Spinal Muscular Atrophy, Type Ii; Spinal Muscular Atrophy, Type Iii; Spinal Muscular Atrophy, Type Iv |
| NCBI Summary: | This gene is part of a 500 kb inverted duplication on chromosome 5q13. This duplicated region contains at least four genes and repetitive elements which make it prone to rearrangements and deletions. The repetitiveness and complexity of the sequence have also caused difficulty in determining the organization of this genomic region. The telomeric and centromeric copies of this gene are nearly identical and encode the same protein. While mutations in the telomeric copy are associated with spinal muscular atrophy, mutations in this gene, the centromeric copy, do not lead to disease. This gene may be a modifier of disease caused by mutation in the telomeric copy. The critical sequence difference between the two genes is a single nucleotide in exon 7, which is thought to be an exon splice enhancer. Note that the nine exons of both the telomeric and centromeric copies are designated historically as exon 1, 2a, 2b, and 3-8. It is thought that gene conversion events may involve the two genes, leading to varying copy numbers of each gene. The full length protein encoded by this gene localizes to both the cytoplasm and the nucleus. Within the nucleus, the protein localizes to subnuclear bodies called gems which are found near coiled bodies containing high concentrations of small ribonucleoproteins (snRNPs). This protein forms heteromeric complexes with proteins such as SIP1 and GEMIN4, and also interacts with several proteins known to be involved in the biogenesis of snRNPs, such as hnRNP U protein and the small nucleolar RNA binding protein. Four transcript variants encoding distinct isoforms have been described. [provided by RefSeq, Sep 2008] |
| UniProt Code: | Q16637 |
| NCBI GenInfo Identifier: | 2498924 |
| NCBI Gene ID: | 6607 |
| NCBI Accession: | Q16637. 1 |
| UniProt Secondary Accession: | Q16637,Q13119, Q549U5, Q96J51, A8K0V4, |
| UniProt Related Accession: | Q16637 |
| Molecular Weight: | 32kDa |
| NCBI Full Name: | Survival motor neuron protein |
| NCBI Synonym Full Names: | survival of motor neuron 2, centromeric |
| NCBI Official Symbol: | SMN2 |
| NCBI Official Synonym Symbols: | SMNC; BCD541; GEMIN1; TDRD16B; C-BCD541 |
| NCBI Protein Information: | survival motor neuron protein |
| UniProt Protein Name: | Survival motor neuron protein |
| UniProt Synonym Protein Names: | Component of gems 1; Gemin-1 |
| UniProt Gene Name: | SMN1 |
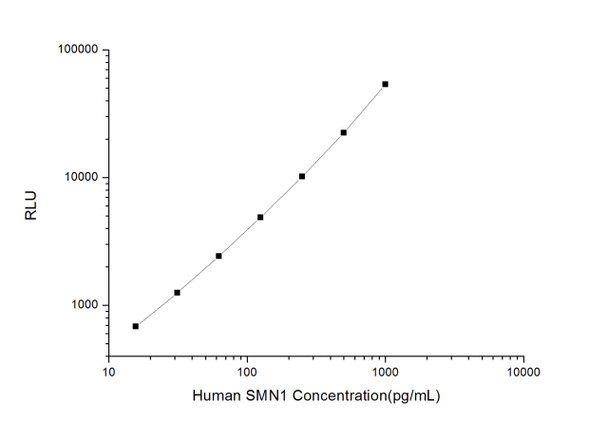
As the RLU values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (pg/mL) | RLU | Average | Corrected |
| 1000 | 51559 55791 | 53675 | 53650 |
| 500 | 20280 24698 | 22489 | 22464 |
| 250 | 10386 10036 | 10211 | 10186 |
| 125 | 4543 5259 | 4901 | 4876 |
| 62.5 | 2641 2265 | 2453 | 2428 |
| 31.25 | 1309 1253 | 1281 | 1256 |
| 15.63 | 696 720 | 708 | 683 |
| 0 | 25 25 | 25 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Human SMN1 were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Human SMN1 were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (pg/mL) | 50.60 | 143.02 | 358.59 | 52.80 | 139.83 | 323.13 |
| Standard deviation | 5.42 | 10.30 | 29.26 | 6.25 | 16.74 | 32.73 |
| C V (%) | 10.71 | 7.20 | 8.16 | 11.84 | 11.97 | 10.13 |
Recovery
The recovery of Human SMN1 spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 97-109 | 103 |
| EDTA plasma (n=5) | 90-104 | 96 |
| Cell culture media (n=5) | 96-110 | 104 |
Linearity
Samples were spiked with high concentrations of Human SMN1 and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 90-103 | 95-110 | 100-115 |
| Average (%) | 95 | 101 | 106 | |
| 1:4 | Range (%) | 100-118 | 93-104 | 102-118 |
| Average (%) | 108 | 98 | 109 | |
| 1:8 | Range (%) | 93-106 | 83-96 | 100-115 |
| Average (%) | 100 | 90 | 109 | |
| 1:16 | Range (%) | 86-100 | 97-112 | 98-115 |
| Average (%) | 93 | 103 | 105 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro CLIA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent A | 1 vial, 5 mL | 4°C (shading light) |
| Substrate Reagent B | 1 vial, 5 mL | 4°C (shading light) |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100 µL of standard solutions into the standard wells.
- Add 100 µL of Sample / Standard dilution buffer into the control (zero) well.
- Add 100 µL of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids. ) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37 °C.
- Aspirate the liquid from each well, do not wash. Immediately add 100 µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37 °C.
- Aspirate or decant the solution from the plate and add 350 µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100 µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37 °C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 100 µL of Substrate mixture solution to each well. Cover with a new plate seal andincubate for no more than 5 min at 37 °C. Protect the plate from light.
- Determine the RLU value of each well immediately.