Human Cell Biology ELISA Kits 6
Human SHH (Hedgehog Homolog, Sonic) ELISA Kit (HUES02054)
- SKU:
- HUES02054
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- Q15465
- Sensitivity:
- 46.88pg/mL
- Range:
- 78.13-5000pg/mL
- ELISA Type:
- Sandwich
- Synonyms:
- HHG1, HLP3, HPE3, MCOPCB5, SMMCI, TPT, TPTPS, Sonic hedgehog
- Reactivity:
- Human
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Cell Biology
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Human |
| Detection Method: | Colormetric |
| Detection Range: | 78.13-5000 pg/mL |
| Sensitivity: | 46.88 pg/mL |
| Sample Volume Required Per Well: | 100µL |
| Sample Type: | Serum, plasma and other biological fluids |
| Specificity: | This kit recognizes Human SHH in samples. No significant cross-reactivity or interference between Human SHH and analogues was observed. |
This ELISA kit uses Sandwich-ELISA as the method. The micro ELISA plate provided in this kit has been pre-coated with an antibody specific to Human SHH. Standards or samples are added to the appropriate micro ELISA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Human SHH and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Human SHH, biotinylated detection antibody and Avidin-HRP conjugate will appear blue in color. The enzyme-substrate reaction is terminated by adding Stop Solution and the color turns yellow. The optical density (OD) is measured spectrophotometrically at a wavelength of 450 nm ± 2 nm. The OD value is proportional to the concentration of Human SHH. The concentration of Human SHH in samples can be calculated by comparing the OD of the samples to the standard curve.
| UniProt Protein Function: | SHH: Binds to the patched (PTC) receptor, which functions in association with smoothened (SMO), to activate the transcription of target genes. In the absence of SHH, PTC represses the constitutive signaling activity of SMO. Also regulates another target, the gli oncogene. Intercellular signal essential for a variety of patterning events during development: signal produced by the notochord that induces ventral cell fate in the neural tube and somites, and the polarizing signal for patterning of the anterior-posterior axis of the developing limb bud. Displays both floor plate- and motor neuron-inducing activity. The threshold concentration of N-product required for motor neuron induction is 5-fold lower than that required for floor plate induction. Interacts with HHATL/GUP1 which negatively regulates HHAT-mediated palmitoylation of the SHH N-terminus. N-product is active as a multimer. Expressed in fetal intestine, liver, lung, and kidney. Not expressed in adult tissues. Belongs to the hedgehog family. |
| UniProt Protein Details: | Protein type:Motility/polarity/chemotaxis; Cell cycle regulation; Oncoprotein; Cell development/differentiation Chromosomal Location of Human Ortholog: 7q36 Cellular Component: extracellular matrix; extracellular space; cell surface; endoplasmic reticulum lumen; plasma membrane; extracellular region; nucleus; cytosol; lipid raft Molecular Function:laminin-1 binding; peptidase activity; morphogen activity; protein binding; glycosaminoglycan binding; zinc ion binding; patched binding; calcium ion binding; glycoprotein binding Biological Process: prostate gland development; central nervous system development; positive regulation of transcription, DNA-dependent; embryonic skeletal development; telencephalon regionalization; embryonic foregut morphogenesis; male genitalia development; neural crest cell migration; inner ear development; embryonic limb morphogenesis; hindbrain development; positive regulation of neuroblast proliferation; camera-type eye development; neuron fate commitment; myotube differentiation; intermediate filament organization; osteoblast development; positive regulation of skeletal muscle cell proliferation; positive regulation of cell division; regulation of proteolysis; positive regulation of transcription from RNA polymerase II promoter; embryonic digit morphogenesis; negative regulation of apoptosis; embryonic forelimb morphogenesis; axon guidance; granule cell precursor proliferation; ventral midline development; spinal cord dorsal/ventral patterning; thalamus development; positive regulation of striated muscle cell differentiation; negative regulation of transcription from RNA polymerase II promoter; palate development; Bergmann glial cell differentiation; negative regulation of T cell proliferation; positive regulation of cell proliferation; pancreas development; forebrain development; thyroid gland development; heart looping; vasculogenesis; negative regulation of cell migration; positive thymic T cell selection; intein-mediated protein splicing; regulation of odontogenesis; androgen metabolic process; spinal cord motor neuron differentiation; pattern specification process; regulation of cell proliferation; odontogenesis of dentine-containing teeth; negative regulation of cell differentiation; stem cell development; embryonic development; dorsal/ventral pattern formation; positive regulation of protein import into nucleus; ureteric bud branching; hindgut morphogenesis; lung development; negative regulation of alpha-beta T cell differentiation; heart development; T cell differentiation in the thymus; CD4-positive or CD8-positive, alpha-beta T cell lineage commitment; lymphoid progenitor cell differentiation; Wnt receptor signaling pathway through beta-catenin; embryonic pattern specification; proteolysis; positive regulation of T cell differentiation in the thymus; cell-cell signaling; embryonic digestive tract morphogenesis; midbrain development; ectoderm development; positive regulation of smoothened signaling pathway; positive regulation of oligodendrocyte differentiation; oligodendrocyte development; activation of hh target transcription factor; striated muscle development; endocytosis; positive regulation of skeletal muscle development; negative thymic T cell selection; patterning of blood vessels; branching morphogenesis of a tube; polarity specification of anterior/posterior axis; metanephros development; cell fate specification; embryonic hindlimb morphogenesis; positive regulation of Wnt receptor signaling pathway; dorsoventral neural tube patterning; smoothened signaling pathway; organ formation; hair follicle morphogenesis; thymus development; smoothened signaling pathway in regulation of granule cell precursor cell proliferation; positive regulation of immature T cell proliferation in the thymus; negative regulation of proteasomal ubiquitin-dependent protein catabolic process; formation of anatomical boundary; myoblast differentiation; limb bud formation; establishment of cell polarity; neuroblast proliferation; blood coagulation; cell development; positive regulation of alpha-beta T cell differentiation Disease: Solitary Median Maxillary Central Incisor; Schizencephaly; Microphthalmia, Isolated, With Coloboma 5; Holoprosencephaly 3 |
| NCBI Summary: | This gene encodes a protein that is instrumental in patterning the early embryo. It has been implicated as the key inductive signal in patterning of the ventral neural tube, the anterior-posterior limb axis, and the ventral somites. Of three human proteins showing sequence and functional similarity to the sonic hedgehog protein of Drosophila, this protein is the most similar. The protein is made as a precursor that is autocatalytically cleaved; the N-terminal portion is soluble and contains the signalling activity while the C-terminal portion is involved in precursor processing. More importantly, the C-terminal product covalently attaches a cholesterol moiety to the N-terminal product, restricting the N-terminal product to the cell surface and preventing it from freely diffusing throughout the developing embryo. Defects in this protein or in its signalling pathway are a cause of holoprosencephaly (HPE), a disorder in which the developing forebrain fails to correctly separate into right and left hemispheres. HPE is manifested by facial deformities. It is also thought that mutations in this gene or in its signalling pathway may be responsible for VACTERL syndrome, which is characterized by vertebral defects, anal atresia, tracheoesophageal fistula with esophageal atresia, radial and renal dysplasia, cardiac anomalies, and limb abnormalities. Additionally, mutations in a long range enhancer located approximately 1 megabase upstream of this gene disrupt limb patterning and can result in preaxial polydactyly. [provided by RefSeq, Jul 2008] |
| UniProt Code: | Q15465 |
| NCBI GenInfo Identifier: | 6094283 |
| NCBI Gene ID: | 6469 |
| NCBI Accession: | Q15465. 1 |
| UniProt Secondary Accession: | Q15465,Q75MC9, A4D247, |
| UniProt Related Accession: | Q15465 |
| Molecular Weight: | 49,607 Da |
| NCBI Full Name: | Sonic hedgehog protein |
| NCBI Synonym Full Names: | sonic hedgehog |
| NCBI Official Symbol: | SHH |
| NCBI Official Synonym Symbols: | TPT; HHG1; HLP3; HPE3; SMMCI; TPTPS; MCOPCB5 |
| NCBI Protein Information: | sonic hedgehog protein; sonic hedgehog homolog |
| UniProt Protein Name: | Sonic hedgehog protein |
| UniProt Synonym Protein Names: | HHG-1Cleaved into the following 2 chains:Sonic hedgehog protein N-product; Sonic hedgehog protein C-product |
| Protein Family: | Sonic hedgehog protein |
| UniProt Gene Name: | SHH |
| UniProt Entry Name: | SHH_HUMAN |
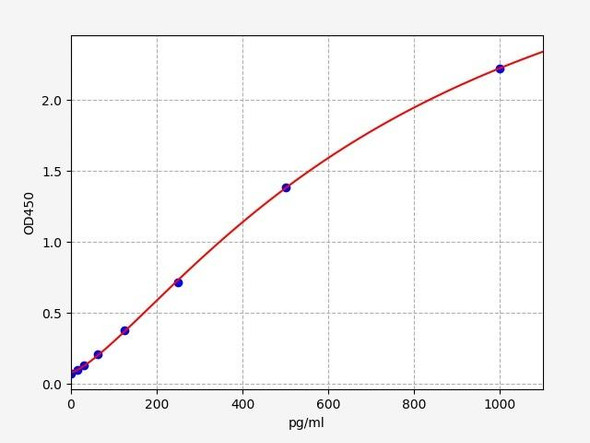
As the OD values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (pg/mL) | O.D | Average | Corrected |
| 5000 | 2.398 2.422 | 2.41 | 2.331 |
| 2500 | 1.753 1.765 | 1.759 | 1.68 |
| 1250 | 1.029 0.997 | 1.013 | 0.934 |
| 625 | 0.509 0.513 | 0.511 | 0.432 |
| 312.5 | 0.276 0.27 | 0.273 | 0.194 |
| 156.25 | 0.194 0.182 | 0.188 | 0.109 |
| 78.13 | 0.131 0.137 | 0.134 | 0.055 |
| 0 | 0.071 0.087 | 0.079 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Human SHH were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Human SHH were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (pg/mL) | 265.23 | 629.21 | 1862.20 | 280.01 | 601.38 | 1709.28 |
| Standard deviation | 13.39 | 32.09 | 59.40 | 15.57 | 26.52 | 68.03 |
| C V (%) | 5.05 | 5.10 | 3.19 | 5.56 | 4.41 | 3.98 |
Recovery
The recovery of Human SHH spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 88-100 | 94 |
| EDTA plasma (n=5) | 94-107 | 101 |
| Cell culture media (n=5) | 87-98 | 92 |
Linearity
Samples were spiked with high concentrations of Human SHH and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 94-110 | 84-97 | 98-110 |
| Average (%) | 102 | 90 | 104 | |
| 1:4 | Range (%) | 87-99 | 83-93 | 81-93 |
| Average (%) | 93 | 88 | 88 | |
| 1:8 | Range (%) | 89-101 | 81-94 | 86-100 |
| Average (%) | 94 | 87 | 93 | |
| 1:16 | Range (%) | 91-104 | 79-90 | 82-92 |
| Average (%) | 97 | 85 | 87 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro ELISA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent | 1 vial, 10 mL | 4°C(shading light) |
| Stop Solution | 1 vial, 10 mL | 4°C |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100 µL of standard solutions into the standard wells.
- Add 100 µL of Sample / Standard dilution buffer into the control (zero) well.
- Add 100 µL of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37 °C.
- Aspirate the liquid from each well, do not wash. Immediately add 100 µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37 °C.
- Aspirate or decant the solution from the plate and add 350 µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100 µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37 °C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 90 µL of Substrate Reagent to each well. Cover with a new plate seal and incubate forapproximately 15 min at 37 °C. Protect the plate from light. Note: the reaction time can beshortened or extended according to the actual color change, but not by more than 30min.
- Add 50 µL of Stop Solution to each well. Note: Adding the stop solution should be done inthe same order as the substrate solution.
- Determine the optical density (OD value) of each well immediately with a microplate readerset at 450 nm.