Human Cardiovascular ELISA Kits
Human PLAU/uPA(Urokinase-Type Plasminogen Activator) CLIA Kit (HUES00991)
- SKU:
- HUES00991
- Product Type:
- ELISA Kit
- ELISA Type:
- CLIA Kit
- Size:
- 96 Assays
- Sensitivity:
- 9.38pg/mL
- Range:
- 15.63-1000pg/mL
- ELISA Type:
- Sandwich
- Synonyms:
- UPA, u-PA, ATF, URK, UP-A, Urokinase, Abbokinase
- Reactivity:
- Human
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Cardiovascular
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Human |
| Detection method: | Chemiluminescence |
| Detection range: | 15.63-1000 pg/mL |
| Sensitivity: | 9.38 pg/mL |
| Sample volume: | 100µL |
| Sample type: | Serum, plasma and other biological fluids |
| Repeatability: | CV < 15% |
| Specificity: | This kit recognizes Human PLAU/uPA in samples. No significant cross-reactivity or interference between Human PLAU/uPA and analogues was observed. |
This kit uses Sandwich-CLIA as the method. The micro CLIA plate provided in this kit has been pre-coated with an antibody specific to Human PLAU/uPA. Standards or samples are added to the appropriate micro CLIA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Human PLAU/uPA and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Human PLAU/uPA, biotinylated detection antibody and Avidin-HRP conjugate will appear fluorescence. The Relative light unit (RLU) value is measured spectrophotometrically by the Chemiluminescence immunoassay analyzer. The RLU value is positively associated with the concentration of Human PLAU/uPA. The concentration of Human PLAU/uPA in the samples can be calculated by comparing the RLU of the samples to the standard curve.
| UniProt Protein Function: | Function: Specifically cleaves the zymogen plasminogen to form the active enzyme plasmin. |
| UniProt Protein Details: | Catalytic activity: Specific cleavage of Arg-|-Val bond in plasminogen to form plasmin. Enzyme regulation: Inhibited by SERPINA5. Ref. 17 Ref. 22 Subunit structure: Found in high and low molecular mass forms. Each consists of two chains, A and B. The high molecular mass form contains a long chain A which is cleaved to yield a short chain A. Forms heterodimer with SERPINA5. Binds LRP1B; binding is followed by internalization and degradation. Interacts with MRC2. Interacts with PLAUR. Ref. 20 Ref. 21 Subcellular location: Secreted. Tissue specificity: Expressed in the prostate gland and prostate cancers. Ref. 23 Post-translational modification: Phosphorylation of Ser-158 and Ser-323 abolishes proadhesive ability but does not interfere with receptor binding. Involvement in Disease: Quebec platelet disorder (QPD) [MIM:601709]: An autosomal dominant bleeding disorder due to a gain-of-function defect in fibrinolysis. Although affected individuals do not exhibit systemic fibrinolysis, they show delayed onset bleeding after challenge, such as surgery. The hallmark of the disorder is markedly increased PLAU levels within platelets, which causes intraplatelet plasmin generation and secondary degradation of alpha-granule proteins. Note: The disease is caused by mutations affecting the gene represented in this entry. Ref. 24 Pharmaceutical use: Available under the name Abbokinase (Abbott). Used in Pulmonary Embolism (PE) to initiate fibrinolysis. Clinically used for therapy of thrombolytic disorders. Sequence similarities: Belongs to the peptidase S1 family. Contains 1 EGF-like domain. Contains 1 kringle domain. Contains 1 peptidase S1 domain. |
| NCBI Summary: | This gene encodes a serine protease involved in degradation of the extracellular matrix and possibly tumor cell migration and proliferation. A specific polymorphism in this gene may be associated with late-onset Alzheimer's disease and also with decreased affinity for fibrin-binding. This protein converts plasminogen to plasmin by specific cleavage of an Arg-Val bond in plasminogen. Plasmin in turn cleaves this protein at a Lys-Ile bond to form a two-chain derivative in which a single disulfide bond connects the amino-terminal A-chain to the catalytically active, carboxy-terminal B-chain. This two-chain derivative is also called HMW-uPA (high molecular weight uPA). HMW-uPA can be further processed into LMW-uPA (low molecular weight uPA) by cleavage of chain A into a short chain A (A1) and an amino-terminal fragment. LMW-uPA is proteolytically active but does not bind to the uPA receptor. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Feb 2009] |
| UniProt Code: | P00749 |
| NCBI GenInfo Identifier: | 254763341 |
| NCBI Gene ID: | 5328 |
| NCBI Accession: | P00749. 2 |
| UniProt Secondary Accession: | P00749,Q15844, Q16618, Q53XS3, Q5SWW9, Q969W6, B4DPZ2 |
| UniProt Related Accession: | P00749 |
| Molecular Weight: | 48,507 Da |
| NCBI Full Name: | Urokinase-type plasminogen activator |
| NCBI Synonym Full Names: | plasminogen activator, urokinase |
| NCBI Official Symbol: | PLAU |
| NCBI Official Synonym Symbols: | ATF; QPD; UPA; URK; u-PA; BDPLT5 |
| NCBI Protein Information: | urokinase-type plasminogen activator; U-plasminogen activator; plasminogen activator, urinary |
| UniProt Protein Name: | Urokinase-type plasminogen activator |
| Protein Family: | Urokinase-type plasminogen activator |
| UniProt Gene Name: | PLAU |
| UniProt Entry Name: | UROK_HUMAN |
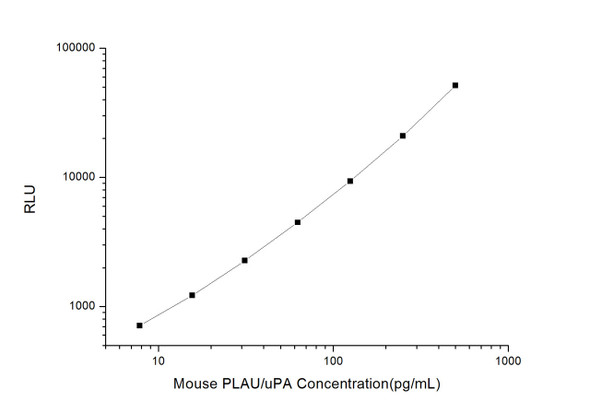
As the RLU values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (pg/mL) | RLU | Average | Corrected |
| 1000 | 52054 55296 | 53675 | 53650 |
| 500 | 20856 24122 | 22489 | 22464 |
| 250 | 11041 9381 | 10211 | 10186 |
| 125 | 4577 5225 | 4901 | 4876 |
| 62.5 | 2591 2315 | 2453 | 2428 |
| 31.25 | 1372 1190 | 1281 | 1256 |
| 15.63 | 696 720 | 708 | 683 |
| 0 | 24 26 | 25 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Human PLAU/uPA were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Human PLAU/uPA were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (pg/mL) | 50.71 | 153.72 | 344.70 | 47.96 | 144.02 | 334.79 |
| Standard deviation | 4.23 | 13.60 | 30.13 | 6.20 | 12.80 | 22.67 |
| C V (%) | 8.34 | 8.85 | 8.74 | 12.93 | 8.89 | 6.77 |
Recovery
The recovery of Human PLAU/uPA spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 98-113 | 105 |
| EDTA plasma (n=5) | 98-112 | 104 |
| Cell culture media (n=5) | 88-103 | 94 |
Linearity
Samples were spiked with high concentrations of Human PLAU/uPA and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 99-113 | 90-104 | 87-100 |
| Average (%) | 106 | 96 | 93 | |
| 1:4 | Range (%) | 93-107 | 99-116 | 86-100 |
| Average (%) | 101 | 106 | 91 | |
| 1:8 | Range (%) | 85-100 | 95-109 | 95-107 |
| Average (%) | 91 | 100 | 102 | |
| 1:16 | Range (%) | 96-111 | 91-104 | 99-115 |
| Average (%) | 102 | 96 | 107 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro CLIA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent A | 1 vial, 5 mL | 4°C (shading light) |
| Substrate Reagent B | 1 vial, 5 mL | 4°C (shading light) |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100 µL of standard solutions into the standard wells.
- Add 100 µL of Sample / Standard dilution buffer into the control (zero) well.
- Add 100 µL of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids. ) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37 °C.
- Aspirate the liquid from each well, do not wash. Immediately add 100 µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37 °C.
- Aspirate or decant the solution from the plate and add 350 µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100 µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37 °C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 100 µL of Substrate mixture solution to each well. Cover with a new plate seal andincubate for no more than 5 min at 37 °C. Protect the plate from light.
- Determine the RLU value of each well immediately.