Human Epigenetics and Nuclear Signaling ELISA Kits
Human PHB (Prohibitin) CLIA Kit (HUES01180)
- SKU:
- HUES01180
- Product Type:
- ELISA Kit
- ELISA Type:
- CLIA Kit
- Size:
- 96 Assays
- Sensitivity:
- 37.5pg/mL
- Range:
- 62.5-4000pg/mL
- ELISA Type:
- Sandwich
- Reactivity:
- Human
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Epigenetics and Nuclear Signaling
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Human |
| Detection method: | Chemiluminescence |
| Detection range: | 62.50-4000 pg/mL |
| Sensitivity: | 37.50 pg/mL |
| Sample volume: | 100µL |
| Sample type: | Serum, plasma and other biological fluids |
| Repeatability: | CV < 15% |
| Specificity: | This kit recognizes Human PHB in samples. No significant cross-reactivity or interference between Human PHB and analogues was observed. |
This kit uses Sandwich-CLIA as the method. The micro CLIA plate provided in this kit has been pre-coated with an antibody specific to Human PHB. Standards or samples are added to the appropriate micro CLIA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Human PHB and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Human PHB, biotinylated detection antibody and Avidin-HRP conjugate will appear fluorescence. The Relative light unit (RLU) value is measured spectrophotometrically by the Chemiluminescence immunoassay analyzer. The RLU value is positively associated with the concentration of Human PHB. The concentration of Human PHB in the samples can be calculated by comparing the RLU of the samples to the standard curve.
| UniProt Protein Function: | PHB: a pleiotropic membrane protein that suppresses cell proliferation, apoptosis and senescence. It plays a role both in maintaining mitochondrial integrity and in cell cycle regulation. Expression increases approximately 3-fold upon entry into G1 phase compared with other phases of the cell cycle. It helps prevent ROS-induced senescence and its expression decreases heterogeneously during cellular aging. Helps maintain the angiogenic capacity of endothelial cells. Up-regulated during activation of primary human T cells via CD3/CD28 pathways. Present in multiple cellular compartments. Large assemblies of PHB and PHB2 localize in the inner membrane of mitochondria. Acts as a mitochondrial chaperone protein, regulates mitochondrial morphogenesis, and helps regulate the organization of mitochondrial DNA. Reported to target lipid rafts. In the nucleus, it has been reported to interact with various transcription factors, modulating their activity. A potential tumor suppressor that functions as a potent transcriptional corepressor for estrogen receptor alpha. It is downregulated by androgens, and represses androgen receptor activity. Co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. In vivo promoter occupancy studies have suggested that the PHB gene is a direct target of c-Myc. May bind to and enhance the transcriptional activity of p53. Both Phb and Phb2 are present in the circulation and can be internalized by cultured cells. Overexpressed in endometrioid ovarian adenocarcinoma and papillary serous ovarian carcinoma. Mutated in sporadic breast cancer. |
| UniProt Protein Details: | Protein type:Oncoprotein; Cell cycle regulation; Mitochondrial; Chaperone; Motility/polarity/chemotaxis; Transcription, coactivator/corepressor; Nuclear receptor co-regulator; Apoptosis Chromosomal Location of Human Ortholog: 17q21 Cellular Component: nucleoplasm; membrane; mitochondrion; integral to plasma membrane; mitochondrial inner membrane; cytoplasm; nucleus Molecular Function:protein binding; enzyme binding; histone deacetylase binding Biological Process: osteoblast differentiation; regulation of apoptosis; transcription from RNA polymerase II promoter; negative regulation of cell proliferation; progesterone receptor signaling pathway; regulation of transcription, DNA-dependent; positive regulation of transcription, DNA-dependent; histone deacetylation; negative regulation of cell growth; negative regulation of transcription from RNA polymerase II promoter; signal transduction; negative regulation of transcription, DNA-dependent Disease: Breast Cancer |
| NCBI Summary: | This gene is evolutionarily conserved, and its product is proposed to play a role in human cellular senescence and tumor suppression. Antiproliferative activity is reported to be localized to the 3' UTR, which is proposed to function as a trans-acting regulatory RNA. Several pseudogenes of this gene have been identified. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013] |
| UniProt Code: | P35232 |
| NCBI GenInfo Identifier: | 464371 |
| NCBI Gene ID: | 5245 |
| NCBI Accession: | P35232. 1 |
| UniProt Secondary Accession: | P35232,Q4VBQ0, B4DY47, |
| UniProt Related Accession: | P35232 |
| Molecular Weight: | 272 |
| NCBI Full Name: | Prohibitin |
| NCBI Synonym Full Names: | prohibitin |
| NCBI Official Symbol: | PHB |
| NCBI Official Synonym Symbols: | PHB1; HEL-215; HEL-S-54e |
| NCBI Protein Information: | prohibitin; epididymis luminal protein 215; epididymis secretory sperm binding protein Li 54e |
| UniProt Protein Name: | Prohibitin |
| Protein Family: | Prohibitin |
| UniProt Gene Name: | PHB |
| UniProt Entry Name: | PHB_HUMAN |
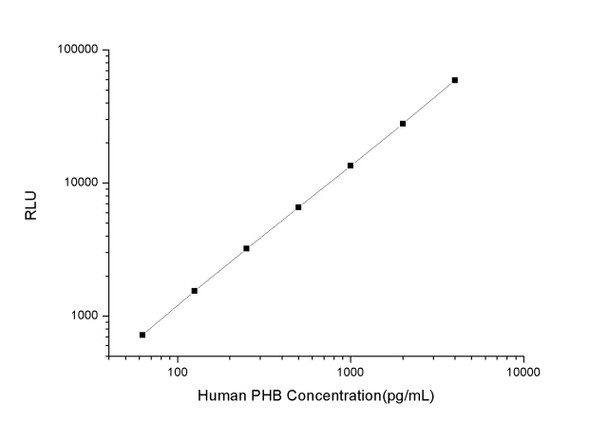
As the RLU values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (pg/mL) | RLU | Average | Corrected |
| 4000 | 56291 61871 | 59081 | 59046 |
| 2000 | 26467 29251 | 27859 | 27824 |
| 1000 | 14030 12938 | 13484 | 13449 |
| 500 | 6049 7161 | 6605 | 6570 |
| 250 | 3301 3185 | 3243 | 3208 |
| 125 | 1685 1477 | 1581 | 1546 |
| 62.50 | 740 770 | 755 | 720 |
| 0 | 35 35 | 35 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Human PHB were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Human PHB were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (pg/mL) | 188.52 | 444.17 | 1380.42 | 185.56 | 469.27 | 1266.23 |
| Standard deviation | 15.27 | 47.35 | 119.68 | 21.32 | 41.58 | 111.55 |
| C V (%) | 8.10 | 10.66 | 8.67 | 11.49 | 8.86 | 8.81 |
Recovery
The recovery of Human PHB spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 91-107 | 98 |
| EDTA plasma (n=5) | 88-102 | 93 |
| Cell culture media (n=5) | 89-104 | 96 |
Linearity
Samples were spiked with high concentrations of Human PHB and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 91-106 | 88-99 | 91-103 |
| Average (%) | 97 | 94 | 98 | |
| 1:4 | Range (%) | 100-111 | 97-111 | 94-108 |
| Average (%) | 105 | 105 | 101 | |
| 1:8 | Range (%) | 96-108 | 89-101 | 100-116 |
| Average (%) | 102 | 96 | 107 | |
| 1:16 | Range (%) | 96-113 | 95-108 | 96-112 |
| Average (%) | 104 | 101 | 103 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro CLIA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent A | 1 vial, 5 mL | 4°C (shading light) |
| Substrate Reagent B | 1 vial, 5 mL | 4°C (shading light) |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100 µL of standard solutions into the standard wells.
- Add 100 µL of Sample / Standard dilution buffer into the control (zero) well.
- Add 100 µL of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids. ) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37 °C.
- Aspirate the liquid from each well, do not wash. Immediately add 100 µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37 °C.
- Aspirate or decant the solution from the plate and add 350 µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100 µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37 °C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 100 µL of Substrate mixture solution to each well. Cover with a new plate seal andincubate for no more than 5 min at 37 °C. Protect the plate from light.
- Determine the RLU value of each well immediately.