Human Epigenetics and Nuclear Signaling ELISA Kits
Human PGR (Progesterone Receptor) ELISA Kit (HUES03154)
- SKU:
- HUES03154
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- P06401
- Sensitivity:
- 0.19ng/mL
- Range:
- 0.31-20ng/mL
- ELISA Type:
- Sandwich
- Reactivity:
- Human
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Epigenetics and Nuclear Signaling
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Human |
| Detection Method: | Colormetric |
| Detection Range: | 0.31-20 ng/mL |
| Sensitivity: | 0.19 ng/mL |
| Sample Volume Required Per Well: | 100µL |
| Sample Type: | Serum, plasma and other biological fluids |
| Specificity: | This kit recognizes Human PGR in samples. No significant cross-reactivity or interference between Human PGR and analogues was observed. |
This ELISA kit uses Sandwich-ELISA as the method. The micro ELISA plate provided in this kit has been pre-coated with an antibody specific to Human PGR. Standards or samples are added to the appropriate micro ELISA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Human PGR and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Human PGR, biotinylated detection antibody and Avidin-HRP conjugate will appear blue in color. The enzyme-substrate reaction is terminated by adding Stop Solution and the color turns yellow. The optical density (OD) is measured spectrophotometrically at a wavelength of 450 nm ± 2 nm. The OD value is proportional to the concentration of Human PGR. The concentration of Human PGR in samples can be calculated by comparing the OD of the samples to the standard curve.
| UniProt Protein Function: | Function: The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Progesterone receptor isoform B (PRB) is involved activation of c-SRC/MAPK signaling on hormone stimulation. Ref. 12 Ref. 20 Ref. 21 Ref. 23 Ref. 24 Ref. 25 Ref. 26 Ref. 27Isoform A:inactive in stimulating c-Src/MAPK signaling on hormone stimulation. Ref. 12 Ref. 20 Ref. 21 Ref. 23 Ref. 24 Ref. 25 Ref. 26 Ref. 27Isoform 4:Increases mitochondrial membrane potential and cellular respiration upon stimulation by progesterone. Ref. 12 Ref. 20 Ref. 21 Ref. 23 Ref. 24 Ref. 25 Ref. 26 Ref. 27 |
| UniProt Protein Details: | Subunit structure: Interacts with SMARD1 and UNC45A. Interacts with CUEDC2; the interaction promotes ubiquitination, decreases sumoylation, and repesses transcriptional activity. Interacts with PIAS3; the interaction promotes sumoylation of PR in a hormone-dependent manner, inhibits DNA-binding, and alters nuclear export. Interacts with SP1; the interaction requires ligand-induced phosphorylation on Ser-345 by ERK1/2 MAPK. Interacts with PRMT2. Ref. 18 Ref. 19 Ref. 22 Ref. 23 Ref. 24 Ref. 27 Subcellular location: Nucleus. Cytoplasm. Note: Nucleoplasmic shuttling is both homone- and cell cycle-dependent. On hormone stimulation, retained in the cytoplasm in the G1 and G2/M phases. Ref. 12 Ref. 20 Ref. 21 Ref. 26Isoform A: Nucleus. Cytoplasm. Note: Mainly nuclear. Ref. 12 Ref. 20 Ref. 21 Ref. 26Isoform 4: Mitochondrion outer membrane Ref. 12 Ref. 20 Ref. 21 Ref. 26. Domain: Composed of three domains: a modulating N-terminal domain, a DNA-binding domain and a C-terminal ligand-binding domain. Post-translational modification: Phosphorylated on multiple serine sites. Several of these sites are hormone-dependent. Phosphorylation on Ser-294 occurs preferentially on isoform B, is highly hormone-dependent and modulates ubiquitination and sumoylation on Lys-388. Phosphorylation on Ser-102 and Ser-345 also requires induction by hormone. Basal phosphorylation on Ser-81, Ser-162, Ser-190 and Ser-400 is increased in response to progesterone and can be phosphorylated in vitro by the CDK2-A1 complex. Increased levels of phosphorylation on Ser-400 also in the presence of EGF, heregulin, IGF, PMA and FBS. Phosphorylation at this site by CDK2 is ligand-independent, and increases nuclear translocation and transcriptional activity. Phosphorylation at Ser-162 and Ser-294, but not at Ser-190, is impaired during the G2/M phase of the cell cycle. Phosphorylation on Ser-345 by ERK1/2 MAPK is required for interaction with SP1. Ref. 11 Ref. 13 Ref. 14 Ref. 15 Ref. 16 Ref. 17 Ref. 20 Ref. 21 Ref. 25 Ref. 26 Ref. 27Sumoylation is hormone-dependent and represses transcriptional activity. Sumoylation on all three sites is enhanced by PIAS3. Desumoylated by SENP1. Sumoylation on Lys-388, the main site of sumoylation, is repressed by ubiquitination on the same site, and modulated by phosphorylation at Ser-294. Ref. 23 Ref. 24 Ref. 25Ubiquitination is hormone-dependent and represses sumoylation on the same site. Promoted by MAPK-mediated phosphorylation on Ser-294. Ref. 23 Ref. 24 Ref. 25Palmitoylated by ZDHHC7 and ZDHHC21. Palmitoylation is required for plasma membrane targeting and for rapid intracellular signaling via ERK and AKT kinases and cAMP generation. Ref. 28 Sequence similarities: Belongs to the nuclear hormone receptor family. NR3 subfamily. Contains 1 nuclear receptor DNA-binding domain. |
| NCBI Summary: | This gene encodes a member of the steroid receptor superfamily. The encoded protein mediates the physiological effects of progesterone, which plays a central role in reproductive events associated with the establishment and maintenance of pregnancy. This gene uses two distinct promotors and translation start sites in the first exon to produce two isoforms, A and B. The two isoforms are identical except for the additional 165 amino acids found in the N-terminus of isoform B and mediate their own response genes and physiologic effects with little overlap. [provided by RefSeq, Jan 2011] |
| UniProt Code: | P06401 |
| NCBI GenInfo Identifier: | 90110048 |
| NCBI Gene ID: | 5241 |
| NCBI Accession: | P06401. 4 |
| UniProt Secondary Accession: | P06401,Q8TDS3, Q9UPF7, A7LQ08, A7X8B0, B4E3T0, |
| UniProt Related Accession: | P06401 |
| Molecular Weight: | |
| NCBI Full Name: | Progesterone receptor |
| NCBI Synonym Full Names: | progesterone receptor |
| NCBI Official Symbol: | PGR |
| NCBI Official Synonym Symbols: | PR; NR3C3 |
| NCBI Protein Information: | progesterone receptor; nuclear receptor subfamily 3 group C member 3 |
| UniProt Protein Name: | Progesterone receptor |
| UniProt Synonym Protein Names: | Nuclear receptor subfamily 3 group C member 3 |
| Protein Family: | Perakine reductase |
| UniProt Gene Name: | PGR |
| UniProt Entry Name: | PRGR_HUMAN |
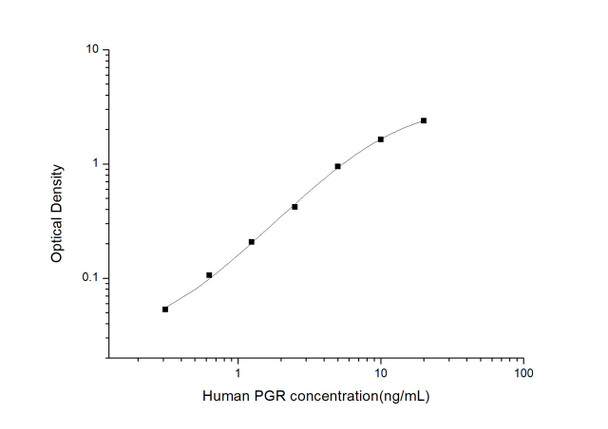
As the OD values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (ng/mL) | O.D | Average | Corrected |
| 20 | 2.419 2.475 | 2.447 | 2.387 |
| 10 | 1.678 1.71 | 1.694 | 1.634 |
| 5 | 1.012 1.008 | 1.01 | 0.95 |
| 2.5 | 0.475 0.485 | 0.48 | 0.42 |
| 1.25 | 0.279 0.255 | 0.267 | 0.207 |
| 0.63 | 0.177 0.155 | 0.166 | 0.106 |
| 0.31 | 0.103 0.123 | 0.113 | 0.053 |
| 0 | 0.057 0.063 | 0.06 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Human PGR were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Human PGR were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (ng/mL) | 0.96 | 2.64 | 7.33 | 0.94 | 2.40 | 7.41 |
| Standard deviation | 0.05 | 0.15 | 0.29 | 0.06 | 0.14 | 0.28 |
| C V (%) | 5.21 | 5.68 | 3.96 | 6.38 | 5.83 | 3.78 |
Recovery
The recovery of Human PGR spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 94-108 | 100 |
| EDTA plasma (n=5) | 92-104 | 99 |
| Cell culture media (n=5) | 92-109 | 100 |
Linearity
Samples were spiked with high concentrations of Human PGR and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 84-99 | 86-99 | 86-99 |
| Average (%) | 91 | 93 | 93 | |
| 1:4 | Range (%) | 90-104 | 88-98 | 87-97 |
| Average (%) | 96 | 93 | 92 | |
| 1:8 | Range (%) | 90-101 | 83-97 | 85-99 |
| Average (%) | 95 | 89 | 91 | |
| 1:16 | Range (%) | 91-107 | 82-94 | 84-97 |
| Average (%) | 98 | 87 | 91 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro ELISA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent | 1 vial, 10 mL | 4°C(shading light) |
| Stop Solution | 1 vial, 10 mL | 4°C |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100µl of standard solutions into the standard wells.
- Add 100µl of Sample / Standard dilution buffer into the control (zero) well.
- Add 100µl of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37°C.
- Aspirate the liquid from each well, do not wash. Immediately add 100µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37°C.
- Aspirate or decant the solution from the plate and add 350µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37°C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 90µL of Substrate Reagent to each well. Cover with a new plate seal and incubate forapproximately 15 min at 37°C. Protect the plate from light. Note: the reaction time can beshortened or extended according to the actual color change, but not by more than 30min.
- Add 50 µL of Stop Solution to each well. Note: Adding the stop solution should be done inthe same order as the substrate solution.
- Determine the optical density (OD value) of each well immediately with a microplate readerset at 450 nm.