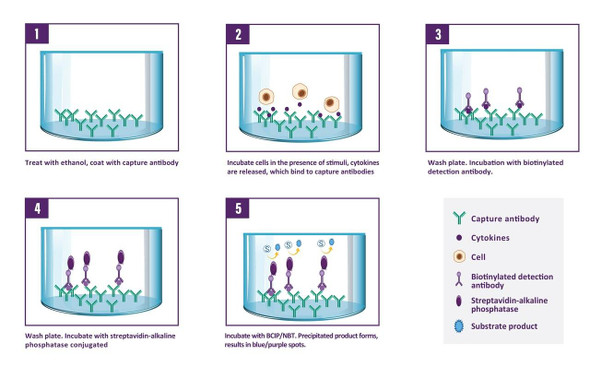
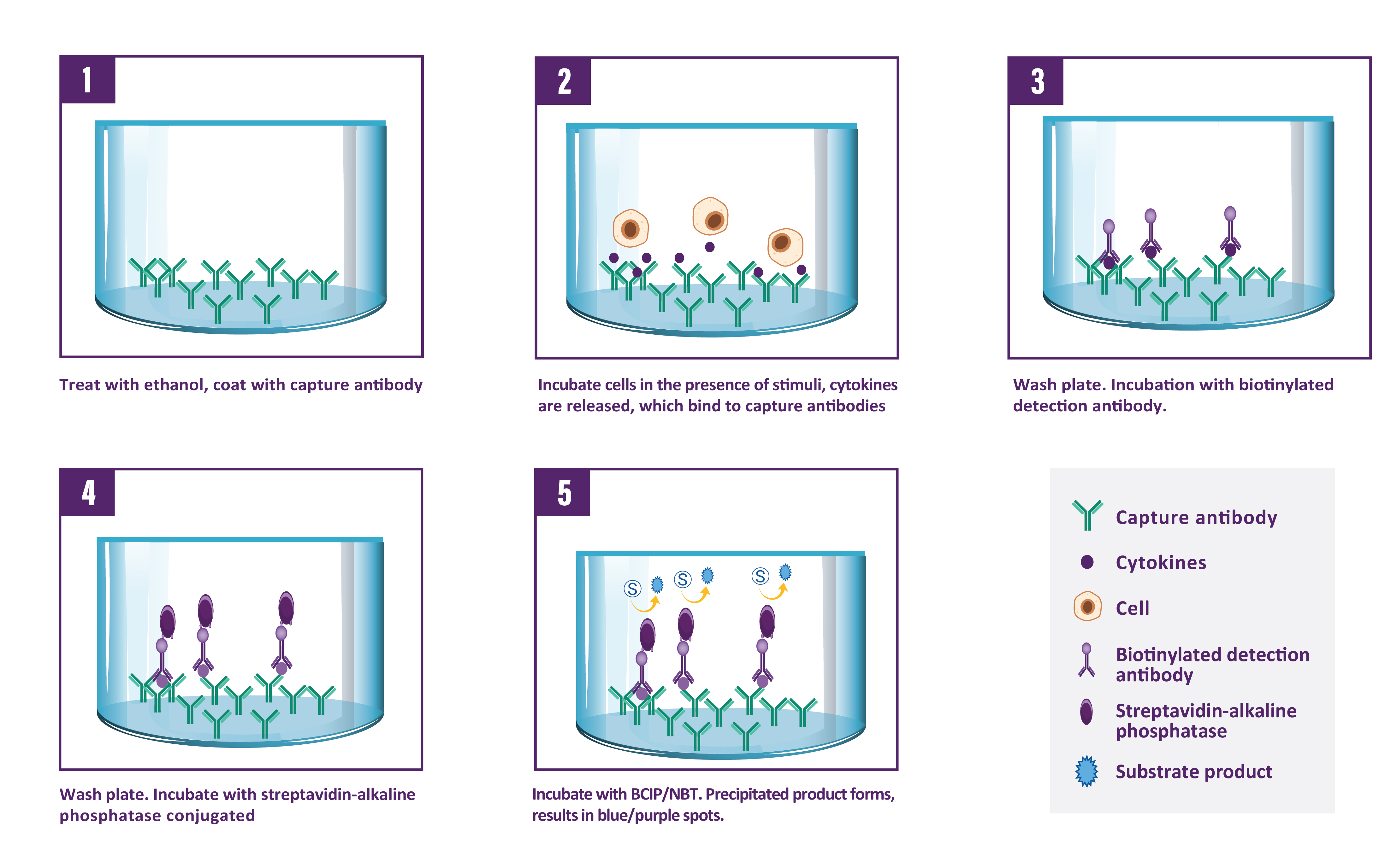
Description
Human Granzyme B ELISpot Kit
Assay Genie ELISpot is a highly specific immunoassay for the analysis of Granzyme B production and secretion from T-cells at a single cell level in conditions closely comparable to the in-vivo environment with minimal cell manipulation. This technique is designed to determine the frequency of Granzyme B producing cells under a given stimulation and the comparison of such frequency against a specific treatment or pathological state. Utilising sandwich immuno-enzyme technology, Assay Genie ELISpot assays can detect both secreted Granzyme B (qualitative analysis) and single cells that produce Granzyme B (quantitative analysis). Cell secreted Granzyme B is captured by coated antibodies avoiding diffusion in supernatant, protease degradation or binding on soluble membrane receptors. After cell removal, the captured Granzyme B is revealed by tracer antibodies and appropriate conjugates.
| Product type: | ELISpot Kit |
| Size: | 10 x 96 Assays |
| Target species: | Human |
| Specificity: | Recognizes natural human Granzyme B |
| Incubation: | 3h after cell stimulation |
| Kit content: | Assay Genie Pre-coated ELISpot kits include precoated PVDF plates, Detection antibody, Alkaline phosphatase conjugate, BSA, BCIP/NBT ready-to-use substrate buffer. |
| Synonyms: | N/A |
| Uniprot: | P10144 |
A capture antibody highly specific for Granzyme B is coated to the wells of a PVDF bottomed 96 well microtitre plate either during kit manufacture or in the laboratory. The plate is then blocked to minimise any non-antibody dependent unspecific binding and washed. Cell suspension and stimulant are added and the plate incubated allowing the specific antibodies to bind any Granzyme B produced. Cells are then removed by washing prior to the addition of Biotinylated detection antibodies which bind to the previously captured Granzyme B. Enzyme conjugated streptavidin is then added binding to the detection antibodies. Following incubation and washing, substrate is then applied to the wells resulting in coloured spots which can be quantified using appropriate analysis software or manually using a microscope.
| Step | Procedure |
| 1. | For PVDF membrane activation, add 25 µl of 35% ethanol to every well. |
| 2. | Incubate plate at room temperature (RT) for 30 seconds. |
| 3. | Empty the wells by flicking the plate over a sink & gently tapping on absorbent paper. Thoroughly wash the plate 3x with 100 µl of PBS 1X per well. |
| 4. | Add 100 µl of diluted capture antibody to every well. |
| 5. | Cover the plate and incubate at 4°C overnight. |
| 6. | Empty the wells as previous and wash the plate once with 100 µl of PBS 1X per well. |
| 7. | Add 100 µl of blocking buffer to every well. |
| 8. | Cover the plate and incubate at RT for 2 hours. |
| 9. | Empty the wells as previous and thoroughly wash once with 100 µl of PBS 1X per well. |
| 10. | Add 100 µl of sample, positive and negative controls cell suspension to appropriate wells providing the required concentration of cells and stimulant. |
| 11. | Cover the plate and incubate at 37°C in a CO2 incubator for an appropriate length of time (15-20 hours). Note: do not agitate or move the plate during this incubation. |
| 12. | Empty the wells and remove excess solution then add 100 µl of Wash Buffer to every well. |
| 13. | Incubate the plate at 4°C for 10 min. |
| 14. | Empty the wells as previous and wash the plate 3x with 100 µl of Wash Buffer. |
| 15. | Add 100 µl of diluted detection antibody to every well. |
| 16. | Cover the plate and incubate at RT for 1 hour 30 min. |
| 17. | Empty the wells as previous and wash the plate 3x with 100 µl of Wash Buffer. |
| 18. | Add 100 µl of diluted Streptavidin-conjugate to every well. |
| 19. | Cover the plate and incubate at RT following the supplier's instructions. |
| 20. | Empty the wells and wash the plate 3x with 100 µl of Wash Buffer. |
| 21. | Peel of the plate bottom and wash both sides of the membrane 3x under running distilled water, once washing complete remove any excess solution by repeated tapping on absorbent paper. |
| 22. | Add 100 µl of ready-to-use substrate buffer to every well. |
| 23. | Following the supplier's instructions, incubate the plate for 5-15 min monitoring spot formation visually throughout the incubation period to assess sufficient colour development. |
| 24. | Empty the wells and rinse both sides of the membrane 3x under running distilled water. Completely remove any excess solution by gentle repeated tapping on absorbent paper Read Spots: allow the wells to dry and then read results. The frequency of the resulting coloured spots. |
| UniProt Protein Function: | GZMB: This enzyme is necessary for target cell lysis in cell- mediated immune responses. It cleaves after Asp. Seems to be linked to an activation cascade of caspases (aspartate-specific cysteine proteases) responsible for apoptosis execution. Cleaves caspase-3, -7, -9 and 10 to give rise to active enzymes mediating apoptosis. By staphylococcal enterotoxin A (SEA) in peripheral blood leukocytes. Inactivated by the serine protease inhibitor diisopropylfluorophosphate. Belongs to the peptidase S1 family. Granzyme subfamily. |
| UniProt Protein Details: | Protein type:Apoptosis; EC 3.4.21.79; Protease Chromosomal Location of Human Ortholog: 14q12 Cellular Component: cytoplasm; cytosol; immunological synapse; intracellular membrane-bound organelle; membrane; nucleus Molecular Function:protein binding; serine-type endopeptidase activity; serine-type peptidase activity Biological Process: apoptosis; natural killer cell mediated cytotoxicity; protein processing |
| NCBI Summary: | This gene encodes a member of the granzyme subfamily of proteins, part of the peptidase S1 family of serine proteases. The encoded preproprotein is secreted by natural killer (NK) cells and cytotoxic T lymphocytes (CTLs) and proteolytically processed to generate the active protease, which induces target cell apoptosis. This protein also processes cytokines and degrades extracellular matrix proteins, and these roles are implicated in chronic inflammation and wound healing. Expression of this gene may be elevated in human patients with cardiac fibrosis. [provided by RefSeq, Sep 2016] |
| UniProt Code: | P10144 |
| NCBI GenInfo Identifier: | 317373361 |
| NCBI Gene ID: | 3002 |
| NCBI Accession: | P10144.2 |
| UniProt Secondary Accession: | P10144,Q8N1D2, Q9UCC1, |
| UniProt Related Accession: | P10144 |
| Molecular Weight: | |
| NCBI Full Name: | Granzyme B |
| NCBI Synonym Full Names: | granzyme B |
| NCBI Official Symbol: | GZMB‚ ‚ |
| NCBI Official Synonym Symbols: | C11; HLP; CCPI; CGL1; CSPB; SECT; CGL-1; CSP-B; CTLA1; CTSGL1‚ ‚ |
| NCBI Protein Information: | granzyme B |
| UniProt Protein Name: | Granzyme B |
| UniProt Synonym Protein Names: | C11; CTLA-1; Cathepsin G-like 1; CTSGL1; Cytotoxic T-lymphocyte proteinase 2; Lymphocyte protease; Fragmentin-2; Granzyme-2; Human lymphocyte protein; HLP; SECT; T-cell serine protease 1-3E |
| Protein Family: | Granzyme |
| UniProt Gene Name: | GZMB‚ ‚ |