Human Metabolism ELISA Kits
Human ApoE (Apolipoprotein E) ELISA Kit (HUES01675)
- SKU:
- HUES01675
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- P02649
- Sensitivity:
- 14.06ng/mL
- Range:
- 23.44-1500ng/mL
- ELISA Type:
- Sandwich
- Synonyms:
- Apo-E, MGC1571, apolipoprotein E3
- Reactivity:
- Human
- Sample Type:
- Serum, plasma and other biological fluids
- Research Area:
- Metabolism
Description
| Assay type: | Sandwich |
| Format: | 96T |
| Assay time: | 4.5h |
| Reactivity: | Human |
| Detection Method: | Colormetric |
| Detection Range: | 23.44-1500 ng/mL |
| Sensitivity: | 14.06 ng/mL |
| Sample Volume Required Per Well: | 100µL |
| Sample Type: | Serum, plasma and other biological fluids |
| Specificity: | This kit recognizes Human ApoE in samples. No significant cross-reactivity or interference between Human ApoE and analogues was observed. |
This ELISA kit uses Sandwich-ELISA as the method. The micro ELISA plate provided in this kit has been pre-coated with an antibody specific to Human ApoE. Standards or samples are added to the appropriate micro ELISA plate wells and combined with the specific antibody. Then a biotinylated detection antibody specific for Human ApoE and Avidin-Horseradish Peroxidase (HRP) conjugate are added to each micro plate well successively and incubated. Free components are washed away. The substrate solution is added to each well. Only those wells that contain Human ApoE, biotinylated detection antibody and Avidin-HRP conjugate will appear blue in color. The enzyme-substrate reaction is terminated by adding Stop Solution and the color turns yellow. The optical density (OD) is measured spectrophotometrically at a wavelength of 450 nm ± 2 nm. The OD value is proportional to the concentration of Human ApoE. The concentration of Human ApoE in samples can be calculated by comparing the OD of the samples to the standard curve.
| UniProt Protein Function: | Function: Mediates the binding, internalization, and catabolism of lipoprotein particles. It can serve as a ligand for the LDL (apo B/E) receptor and for the specific apo-E receptor (chylomicron remnant) of hepatic tissues. |
| UniProt Protein Details: | Subcellular location: Secreted. Tissue specificity: Occurs in all lipoprotein fractions in plasma. It constitutes 10-20% of very low density lipoproteins (VLDL) and 1-2% of high density lipoproteins (HDL). APOE is produced in most organs. Significant quantities are produced in liver, brain, spleen, lung, adrenal, ovary, kidney and muscle. Ref. 18 Post-translational modification: Synthesized with the sialic acid attached by O-glycosidic linkage and is subsequently desialylated in plasma. O-glycosylated with core 1 or possibly core 8 glycans. Thr-307 is a minor glycosylation site compared to Ser-308. Ref. 19Glycated in plasma VLDL of normal subjects, and of hyperglycemic diabetic patients at a higher level (2-3 fold). Phosphorylation sites are present in the extracelllular medium. Polymorphism: Three common APOE alleles have been identified: APOE*2, APOE*3, and APOE*4. The corresponding three major isoforms, E2, E3, and E4, are recognized according to their relative position after isoelectric focusing. Different mutations causing the same migration pattern after isoelectric focusing define different isoform subtypes. The most common isoform is E3 and is present in 40-90% of the population. Common APOE variants influence lipoprotein metabolism in healthy individuals. Involvement in Disease: Defects in APOE are a cause of hyperlipoproteinemia type 3 (HLPP3) [ MIM:107741]; also known as familial dysbetalipoproteinemia. Individuals with HLPP3 are clinically characterized by xanthomas, yellowish lipid deposits in the palmar crease, or less specific on tendons and on elbows. The disorder rarely manifests before the third decade in men. In women, it is usually expressed only after the menopause. The vast majority of the patients are homozygous for APOE*2 alleles. More severe cases of HLPP3 have also been observed in individuals heterozygous for rare APOE variants. The influence of APOE on lipid levels is often suggested to have major implications for the risk of coronary artery disease (CAD). Individuals carrying the common APOE*4 variant are at higher risk of CAD. Ref. 16 Ref. 26 Ref. 27 Ref. 29Genetic variations in APOE are associated with Alzheimer disease type 2 (AD2) [ MIM:104310]. It is a late-onset neurodegenerative disorder characterized by progressive dementia, loss of cognitive abilities, and deposition of fibrillar amyloid proteins as intraneuronal neurofibrillary tangles, extracellular amyloid plaques and vascular amyloid deposits. The major constituent of these plaques is the neurotoxic amyloid-beta-APP 40-42 peptide (s), derived proteolytically from the transmembrane precursor protein APP by sequential secretase processing. The cytotoxic C-terminal fragments (CTFs) and the caspase-cleaved products such as C31 derived from APP, are also implicated in neuronal death. Note=The APOE*4 allele is genetically associated with the common late onset familial and sporadic forms of Alzheimer disease. Risk for AD increased from 20% to 90% and mean age at onset decreased from 84 to 68 years with increasing number of APOE*4 alleles in 42 families with late onset AD. Thus APOE*4 gene dose is a major risk factor for late onset AD and, in these families, homozygosity for APOE*4 was virtually sufficient to cause AD by age 80. The mechanism by which APOE*4 participates in pathogenesis is not known. Ref. 16Defects in APOE are a cause of sea-blue histiocyte disease (SBHD) [ MIM:269600]; also known as sea-blue histiocytosis. This disorder is characterized by splenomegaly, mild thrombocytopenia and, in the bone marrow, numerous histiocytes containing cytoplasmic granules which stain bright blue with the usual hematologic stains. The syndrome is the consequence of an inherited metabolic defect analogous to Gaucher disease and other sphingolipidoses. Ref. 16 Ref. 33 Ref. 36Defects in APOE are a cause of lipoprotein glomerulopathy (LPG) [ MIM:611771]. LPG is an uncommon kidney disease characterized by proteinuria, progressive kidney failure, and distinctive lipoprotein thrombi in glomerular capillaries. It mainly affects people of Japanese and Chinese origin. The disorder has rarely been described in Caucasians. Ref. 16 Ref. 30 Ref. 32 Ref. 37 Sequence similarities: Belongs to the apolipoprotein A1/A4/E family. |
| NCBI Summary: | Chylomicron remnants and very low density lipoprotein (VLDL) remnants are rapidly removed from the circulation by receptor-mediated endocytosis in the liver. Apolipoprotein E, a main apoprotein of the chylomicron, binds to a specific receptor on liver cells and peripheral cells. ApoE is essential for the normal catabolism of triglyceride-rich lipoprotein constituents. The APOE gene is mapped to chromosome 19 in a cluster with APOC1 and APOC2. Defects in apolipoprotein E result in familial dysbetalipoproteinemia, or type III hyperlipoproteinemia (HLP III), in which increased plasma cholesterol and triglycerides are the consequence of impaired clearance of chylomicron and VLDL remnants. [provided by RefSeq] |
| UniProt Code: | P02649 |
| NCBI GenInfo Identifier: | 114039 |
| NCBI Gene ID: | 348 |
| NCBI Accession: | P02649. 1 |
| UniProt Secondary Accession: | P02649,Q9P2S4, B2RC15, C0JYY5, |
| UniProt Related Accession: | P02649,Q13791,Q6LA97,Q6LBZ1,Q8TCZ8 |
| Molecular Weight: | |
| NCBI Full Name: | Apolipoprotein E |
| NCBI Synonym Full Names: | apolipoprotein E |
| NCBI Official Symbol: | APOE |
| NCBI Official Synonym Symbols: | AD2; LPG; LDLCQ5; MGC1571 |
| NCBI Protein Information: | apolipoprotein E; apo-E; apolipoprotein E3; OTTHUMP00000159143; OTTHUMP00000197075; OTTHUMP00000197076; OTTHUMP00000197077 |
| UniProt Protein Name: | Apolipoprotein E |
| Protein Family: | Apolipoprotein |
| UniProt Gene Name: | APOE |
| UniProt Entry Name: | APOE_HUMAN |
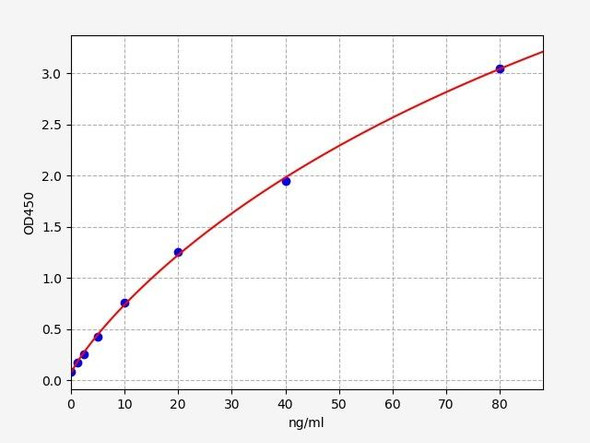
As the OD values of the standard curve may vary according to the conditions of the actual assay performance (e. g. operator, pipetting technique, washing technique or temperature effects), the operator should establish a standard curve for each test. Typical standard curve and data is provided below for reference only.
| Concentration (ng/mL) | O.D | Average | Corrected |
| 1500 | 2.373 2.427 | 2.4 | 2.332 |
| 750 | 1.719 1.731 | 1.725 | 1.657 |
| 375 | 0.975 0.949 | 0.962 | 0.894 |
| 187.5 | 0.442 0.462 | 0.452 | 0.384 |
| 93.75 | 0.261 0.241 | 0.251 | 0.183 |
| 46.88 | 0.183 0.163 | 0.173 | 0.105 |
| 23.44 | 0.115 0.129 | 0.122 | 0.054 |
| 0 | 0.062 0.074 | 0.068 | -- |
Precision
Intra-assay Precision (Precision within an assay): 3 samples with low, mid range and high level Human ApoE were tested 20 times on one plate, respectively.
Inter-assay Precision (Precision between assays): 3 samples with low, mid range and high level Human ApoE were tested on 3 different plates, 20 replicates in each plate.
| Intra-assay Precision | Inter-assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean (ng/mL) | 80.40 | 156.20 | 548.40 | 77.00 | 169.10 | 518.60 |
| Standard deviation | 5.00 | 7.30 | 25.80 | 5.20 | 7.80 | 28.00 |
| C V (%) | 6.22 | 4.67 | 4.70 | 6.75 | 4.61 | 5.40 |
Recovery
The recovery of Human ApoE spiked at three different levels in samples throughout the range of the assay was evaluated in various matrices.
| Sample Type | Range (%) | Average Recovery (%) |
| Serum (n=5) | 93-109 | 101 |
| EDTA plasma (n=5) | 95-107 | 102 |
| Cell culture media (n=5) | 90-105 | 97 |
Linearity
Samples were spiked with high concentrations of Human ApoE and diluted with Reference Standard & Sample Diluent to produce samples with values within the range of the assay.
| Serum (n=5) | EDTA plasma (n=5) | Cell culture media (n=5) | ||
| 1:2 | Range (%) | 84-98 | 97-110 | 89-105 |
| Average (%) | 90 | 103 | 97 | |
| 1:4 | Range (%) | 97-112 | 87-100 | 91-107 |
| Average (%) | 103 | 92 | 98 | |
| 1:8 | Range (%) | 95-112 | 87-102 | 97-109 |
| Average (%) | 102 | 93 | 102 | |
| 1:16 | Range (%) | 101-113 | 83-95 | 93-105 |
| Average (%) | 107 | 90 | 99 |
An unopened kit can be stored at 4°C for 1 month. If the kit is not used within 1 month, store the items separately according to the following conditions once the kit is received.
| Item | Specifications | Storage |
| Micro ELISA Plate(Dismountable) | 8 wells ×12 strips | -20°C, 6 months |
| Reference Standard | 2 vials | |
| Concentrated Biotinylated Detection Ab (100×) | 1 vial, 120 µL | |
| Concentrated HRP Conjugate (100×) | 1 vial, 120 µL | -20°C(shading light), 6 months |
| Reference Standard & Sample Diluent | 1 vial, 20 mL | 4°C, 6 months |
| Biotinylated Detection Ab Diluent | 1 vial, 14 mL | |
| HRP Conjugate Diluent | 1 vial, 14 mL | |
| Concentrated Wash Buffer (25×) | 1 vial, 30 mL | |
| Substrate Reagent | 1 vial, 10 mL | 4°C(shading light) |
| Stop Solution | 1 vial, 10 mL | 4°C |
| Plate Sealer | 5 pieces | |
| Product Description | 1 copy | |
| Certificate of Analysis | 1 copy |
- Set standard, test sample and control (zero) wells on the pre-coated plate and record theirpositions. It is recommended to measure each standard and sample in duplicate. Note: addall solutions to the bottom of the plate wells while avoiding contact with the well walls. Ensuresolutions do not foam when adding to the wells.
- Aliquot 100 µL of standard solutions into the standard wells.
- Add 100 µL of Sample / Standard dilution buffer into the control (zero) well.
- Add 100 µL of properly diluted sample (serum, plasma, tissue homogenates and otherbiological fluids) into test sample wells.
- Cover the plate with the sealer provided in the kit and incubate for 90 min at 37 °C.
- Aspirate the liquid from each well, do not wash. Immediately add 100 µL of BiotinylatedDetection Ab working solution to each well. Cover the plate with a plate seal and gently mix. Incubate for 1 hour at 37 °C.
- Aspirate or decant the solution from the plate and add 350 µL of wash buffer to each welland incubate for 1-2 minutes at room temperature. Aspirate the solution from each well andclap the plate on absorbent filter paper to dry. Repeat this process 3 times. Note: a microplatewasher can be used in this step and other wash steps.
- Add 100 µL of HRP Conjugate working solution to each well. Cover with a plate seal andincubate for 30 min at 37 °C.
- Aspirate or decant the solution from each well. Repeat the wash process for five times asconducted in step 7.
- Add 90 µL of Substrate Reagent to each well. Cover with a new plate seal and incubate forapproximately 15 min at 37 °C. Protect the plate from light. Note: the reaction time can beshortened or extended according to the actual color change, but not by more than 30min.
- Add 50 µL of Stop Solution to each well. Note: Adding the stop solution should be done inthe same order as the substrate solution.
- Determine the optical density (OD value) of each well immediately with a microplate readerset at 450 nm.