Description
| Antibody Name: | hns Antibody (PACO63619) |
| Antibody SKU: | PACO63619 |
| Size: | 50ul |
| Host Species: | Rabbit |
| Tested Applications: | ELISA, WB |
| Recommended Dilutions: | ELISA:1:2000-1:10000, WB:1:2000-1:10000 |
| Species Reactivity: | Escherichia coli |
| Immunogen: | Recombinant Escherichia coli DNA-binding protein H-NS protein (39-137AA) |
| Form: | Liquid |
| Storage Buffer: | Preservative: 0.03% Proclin 300 Constituents: 50% Glycerol, 0.01M PBS, pH 7.4 |
| Purification Method: | >95%, Protein G purified |
| Clonality: | Polyclonal |
| Isotype: | IgG |
| Conjugate: | Non-conjugated |
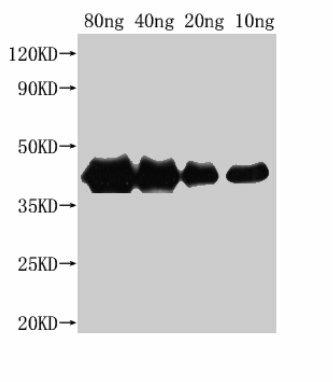 | Western Blot. Positive WB detected in Recombinant protein. All lanes: hns antibody at 1:2500. Secondary. Goat polyclonal to rabbit IgG at 1/50000 dilution. Predicted band size: 38 kDa. Observed band size: 40 kDa. |
| Background: | A DNA-binding protein implicated in transcriptional repression (silencing). Also involved in bacterial chromosome organization and compaction. H-NS binds tightly to AT-rich dsDNA and inhibits transcription. Binds upstream and downstream of initiating RNA polymerase, trapping it in a loop and preventing transcription. Binds to hundreds of sites, approximately half its binding sites are in non-coding DNA, which only accounts for about 10% of the genome. Many of these loci were horizontally transferred (HTG); this offers the selective advantage of silencing foreign DNA while keeping it in the genome in case of need. Suppresses transcription at many intragenic sites as well as transcription of spurious, non-coding RNAs genome-wide. Repression of HTG by H-NS is thought to allow their DNA to evolve faster than non-H-NS-bound regions, and facilitates integration of HTG into transcriptional regulatory networks. A subset of H-NSStpA-regulated genes also require Hha (andor Cnu, ydgT) for repression; Hha and Cnu increase the number of genes DNA bound by H-NSStpA and may also modulate the oligomerization of the H-NSStpA-complex. The protein forms 2 clusters in the nucleoid which gather hns-bound loci together, bridging non-contiguous DNA, and causes DNA substantial condensation. Binds DNA better at low temperatures than at 37 degrees Celsius; AT-rich sites nucleate H-NS binding, further DNA-binding is cooperative and this cooperativity decreases with rising temperature. Transcriptional repression can be inhibited by dominant-negative mutants of StpA or itself. May effect transcriptional elongation. Can increase translational efficiency of mRNA with suboptimal Shine-Dalgarno sequences. Plays a role in the thermal control of pili and adhesive curli fimbriae production, by inducing transcription of csgD. Plays a role in flagellar function. Represses the CRISPR-cas promoters, permits only weak transcription of the crRNA precursor; its repression is antagonized by LeuO. Binds preferentially to the upstream region of its own gene recognizing two segments of DNA on both sides of a bend centered around -150. Overexpression suppresses secY24, a temperature-sensitive mutation. Has also been reported to activate transcription of some genes. |
| Synonyms: | DNA-binding protein H-NS, Heat-stable nucleoid-structuring protein, Histone-like protein HLP-II, Protein B1, Protein H1, hns, bglY, cur, drdX, hnsA, msyA, osmZ, pilG, topS |
| UniProt Protein Function: | A DNA-binding protein implicated in transcriptional repression (silencing) (PubMed:333393, PubMed:2128918, PubMed:8890170, PubMed:8913298, PubMed:9398522, PubMed:16963779, PubMed:17046956, PubMed:23543115). Also involved in bacterial chromosome organization and compaction (PubMed:6379600, PubMed:10982869, PubMed:21903814). H-NS binds tightly to AT-rich dsDNA and inhibits transcription (PubMed:2512122, PubMed:16963779, PubMed:17435766, PubMed:17881364, PubMed:23543115). Binds upstream and downstream of initiating RNA polymerase, trapping it in a loop and preventing transcription (PubMed:11714691). Binds to hundreds of sites, approximately half its binding sites are in non-coding DNA, which only accounts for about 10% of the genome (PubMed:16963779, PubMed:17046956, PubMed:23543115). Many of these loci were horizontally transferred (HTG); this offers the selective advantage of silencing foreign DNA while keeping it in the genome in case of need (PubMed:17046956, PubMed:17881364, PubMed:26789284). Suppresses transcription at many intragenic sites as well as transcription of spurious, non-coding RNAs genome-wide (PubMed:24449106). Repression of HTG by H-NS is thought to allow their DNA to evolve faster than non-H-NS-bound regions, and facilitates integration of HTG into transcriptional regulatory networks (PubMed:26789284). A subset of H-NS/StpA-regulated genes also require Hha (and/or Cnu, ydgT) for repression; Hha and Cnu increase the number of genes DNA bound by H-NS/StpA and may also modulate the oligomerization of the H-NS/StpA-complex (PubMed:23543115). The protein forms 2 clusters in the nucleoid which gather hns-bound loci together, bridging non-contiguous DNA, and causes DNA substantial condensation (PubMed:21903814). Binds DNA better at low temperatures than at 37 degrees Celsius; AT-rich sites nucleate H-NS binding, further DNA-binding is cooperative and this cooperativity decreases with rising temperature (PubMed:17435766, PubMed:17881364). Transcriptional repression can be inhibited by dominant-negative mutants of StpA or itself (PubMed:8755860). May effect transcriptional elongation (PubMed:25638302). Can increase translational efficiency of mRNA with suboptimal Shine-Dalgarno sequences (PubMed:20595230). Plays a role in the thermal control of pili and adhesive curli fimbriae production, by inducing transcription of csgD (PubMed:17010156). Plays a role in flagellar function (PubMed:11031114). Represses the CRISPR-cas promoters, permits only weak transcription of the crRNA precursor; its repression is antagonized by LeuO (PubMed:20132443, PubMed:20659289). Binds preferentially to the upstream region of its own gene recognizing two segments of DNA on both sides of a bend centered around -150 (PubMed:7934818). Overexpression suppresses secY24, a temperature-sensitive mutation (PubMed:1537791). Has also been reported to activate transcription of some genes (PubMed:4566454, PubMed:338303, PubMed:2128918). |
| UniProt Protein Details: | |
| NCBI Summary: | HN-S forms heteromeric complexes with Hha and Cnu, which modulates HN-S function, expression and stability. [More information is available at EcoGene: EG10457]. Heat-stable nucleoid-structuring protein (H-NS) is a nucleoid-associated protein that acts as a transcriptional repressor and may serve to generally suppress horizontally acquired genes. [More information is available at EcoCyc: EG10457]. |
| UniProt Code: | P0ACF8 |
| NCBI GenInfo Identifier: | 16129198 |
| NCBI Gene ID: | 945829 |
| NCBI Accession: | NP_415753.1 |
| UniProt Secondary Accession: | P0ACF8,P08936, Q47267 |
| UniProt Related Accession: | P0ACF8 |
| Molecular Weight: | 19.5 kDa |
| NCBI Full Name: | global DNA-binding transcriptional dual regulator H-NS |
| NCBI Synonym Full Names: | |
| NCBI Official Symbol: | hns |
| NCBI Official Synonym Symbols: | B1; bglY; cur; drc; drdX; drs; ECK1232; fimG; H1; hnsA; irk; JW1225; msyA; osmZ; pilG; topS; topX; verA; virR |
| NCBI Protein Information: | global DNA-binding transcriptional dual regulator H-NS |
| UniProt Protein Name: | DNA-binding protein H-NS |
| UniProt Synonym Protein Names: | Heat-stable nucleoid-structuring protein |
| Protein Family: | DNA-binding protein |
| UniProt Gene Name: | hns |
| UniProt Entry Name: |






