Epigenetics & Nuclear Signaling Antibodies 5
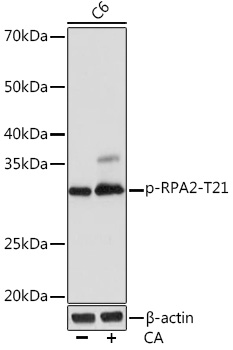
Anti-Phospho-RPA2-T21 Antibody (CABP1040)
- SKU:
- CABP1040
- Product Type:
- Antibody
- Reactivity:
- Human
- Reactivity:
- Rat
- Host Species:
- Rabbit
- Isotype:
- IgG
- Antibody Type:
- Monoclonal Antibody
- Research Area:
- Epigenetics and Nuclear Signaling
Description
| Antibody Name: | Anti-Phospho-RPA2-T21 Antibody |
| Antibody SKU: | CABP1040 |
| Antibody Size: | 20uL, 50uL, 100uL |
| Application: | WB |
| Reactivity: | Human, Rat |
| Host Species: | Rabbit |
| Immunogen: | A phospho specific peptide corresponding to residues surrounding T21 of human RPA2 |
| Application: | WB |
| Recommended Dilution: | WB 1:500 - 1:2000 |
| Reactivity: | Human, Rat |
| Positive Samples: | C6 |
| Immunogen: | A phospho specific peptide corresponding to residues surrounding T21 of human RPA2 |
| Purification Method: | Affinity purification |
| Storage Buffer: | Store at -20°C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 0.05% BSA, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | Email for sequence |
| Gene ID: | 6118 |
| Uniprot: | P15927 |
| Cellular Location: | |
| Calculated MW: | 32kDa |
| Observed MW: | 32KDa |
| Synonyms: | REPA2, RP-A p32, RP-A p34, RPA32 |
| Background: | As part of the heterotrimeric replication protein A complex (RPA/RP-A), binds and stabilizes single-stranded DNA intermediates, that form during DNA replication or upon DNA stress. It prevents their reannealing and in parallel, recruits and activates different proteins and complexes involved in DNA metabolism. Thereby, it plays an essential role both in DNA replication and the cellular response to DNA damage. In the cellular response to DNA damage, the RPA complex controls DNA repair and DNA damage checkpoint activation. Through recruitment of ATRIP activates the ATR kinase a master regulator of the DNA damage response. It is required for the recruitment of the DNA double-strand break repair factors RAD51 and RAD52 to chromatin in response to DNA damage. Also recruits to sites of DNA damage proteins like XPA and XPG that are involved in nucleotide excision repair and is required for this mechanism of DNA repair. Plays also a role in base excision repair (BER) probably through interaction with UNG. Also recruits SMARCAL1/HARP, which is involved in replication fork restart, to sites of DNA damage. May also play a role in telomere maintenance. |
| UniProt Protein Function: | RFA2: Required for DNA recombination, repair and replication. The activity of RP-A is mediated by single-stranded DNA binding and protein interactions. Required for the efficient recruitment of the DNA double-strand break repair factor RAD51 to chromatin in response to DNA damage. Heterotrimer of 70, 32 and 14 kDa chains (canonical replication protein A complex). Component of the alternative replication protein A complex (aRPA) composed of RPA1, RPA3 and RPA4. The DNA-binding activity may reside exclusively on the 70 kDa subunit. Binds to SERTAD3/RBT1. Interacts with TIPIN. Directly interacts with PPP4R2, but not with SMEK2; this interaction is DNA damage-dependent and leads RPA2 dephosphorylation by PPP4C recruitment. Interacts with RAD51, preferentially when hyperphosphorylated. Directly interacts with RFWD3. 3 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:DNA replication Chromosomal Location of Human Ortholog: 1p35 Cellular Component: chromatin; chromosome, telomeric region; DNA replication factor A complex; nuclear chromosome, telomeric region; nucleoplasm; nucleus; PML body Molecular Function:damaged DNA binding; enzyme binding; protein binding; protein N-terminus binding; protein phosphatase binding; single-stranded DNA binding; ubiquitin protein ligase binding Biological Process: base-excision repair; bypass DNA synthesis; DNA damage response, detection of DNA damage; DNA repair; DNA replication; DNA strand elongation during DNA replication; double-strand break repair; double-strand break repair via homologous recombination; error-prone postreplication DNA repair; G1 DNA damage checkpoint; G1/S transition of mitotic cell cycle; mismatch repair; mitotic cell cycle; nucleotide-excision repair; nucleotide-excision repair, DNA gap filling; nucleotide-excision repair, DNA incision; telomere maintenance; telomere maintenance via recombination; telomere maintenance via semi-conservative replication; transcription-coupled nucleotide-excision repair |
| UniProt Code: | P15927 |
| NCBI GenInfo Identifier: | 132474 |
| NCBI Gene ID: | 6118 |
| NCBI Accession: | P15927.1 |
| UniProt Secondary Accession: | P15927,Q52II0, Q5TEI9, Q5TEJ5, |
| UniProt Related Accession: | P15927 |
| Molecular Weight: | 38,810 Da |
| NCBI Full Name: | Replication protein A 32 kDa subunit |
| NCBI Synonym Full Names: | replication protein A2 |
| NCBI Official Symbol: | RPA2 |
| NCBI Official Synonym Symbols: | REPA2; RPA32; RP-A p32; RP-A p34 |
| NCBI Protein Information: | replication protein A 32 kDa subunit |
| UniProt Protein Name: | Replication protein A 32 kDa subunit |
| UniProt Synonym Protein Names: | Replication factor A protein 2; RF-A protein 2; Replication protein A 34 kDa subunit; RP-A p34 |
| Protein Family: | Replication protein |
| UniProt Gene Name: | RPA2 |
| UniProt Entry Name: | RFA2_HUMAN |