Epigenetics & Nuclear Signaling Antibodies 1
Anti-OGG1 Antibody (CAB1384)
- SKU:
- CAB1384
- Product Type:
- Antibody
- Reactivity:
- Human
- Reactivity:
- Mouse
- Reactivity:
- Rat
- Host Species:
- Rabbit
- Isotype:
- IgG
- Antibody Type:
- Polyclonal Antibody
- Research Area:
- Epigenetics and Nuclear Signaling
Description
| Antibody Name: | Anti-OGG1 Antibody |
| Antibody SKU: | CAB1384 |
| Antibody Size: | 20uL, 50uL, 100uL |
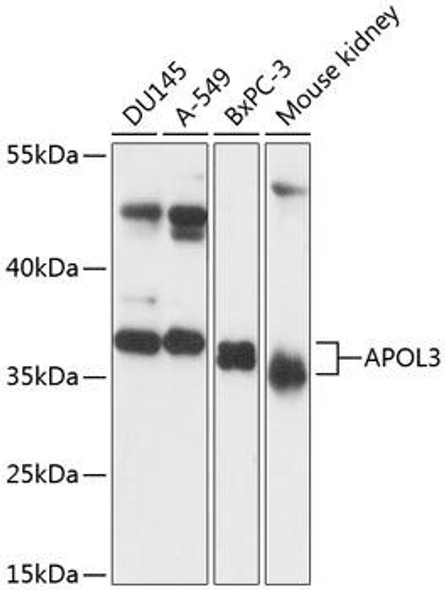
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Host Species: | Rabbit |
| Immunogen: | Recombinant fusion protein containing a sequence corresponding to amino acids 1-345 of human OGG1 (NP_002533.1). |
| Application: | WB IHC |
| Recommended Dilution: | WB 1:500 - 1:2000 IHC 1:50 - 1:200 |
| Reactivity: | Human, Mouse, Rat |
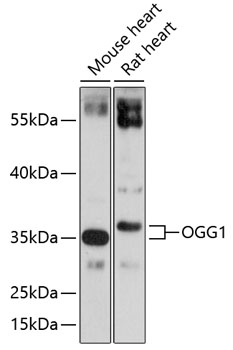
| Positive Samples: | Mouse heart, Rat heart |
| Immunogen: | Recombinant fusion protein containing a sequence corresponding to amino acids 1-345 of human OGG1 (NP_002533.1). |
| Purification Method: | Affinity purification |
| Storage Buffer: | Store at -20'C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | MPAR ALLP RRMG HRTL ASTP ALWA SIPC PRSE LRLD LVLP SGQS FRWR EQSP AHWS GVLA DQVW TLTQ TEEQ LHCT VYRG DKSQ ASRP TPDE LEAV RKYF QLDV TLAQ LYHH WGSV DSHF QEVA QKFQ GVRL LRQD PIEC LFSF ICSS NNNI ARIT GMVE RLCQ AFGP RLIQ LDDV TYHG FPSL QALA GPEV EAHL RKLG LGYR ARYV SASA RAIL EEQG GLAW LQQL RESS YEEA HKAL CILP GVGT KVAD CICL MALD KPQA VPVD VHMW HIAQ RDYS WHPT TSQA KGPS PQTN KELG NFFR SLWG PYAG WAQA VLFS ADLR QSRH AQEP PAKR RKGS KGPE G |
| Gene ID: | 4968 |
| Uniprot: | O15527 |
| Cellular Location: | Mitochondrion, Nucleus, Nucleus matrix, Nucleus speckle, nucleoplasm |
| Calculated MW: | 22kDa/36kDa/38kDa/39kDa/40kDa/45kDa/47kDa |
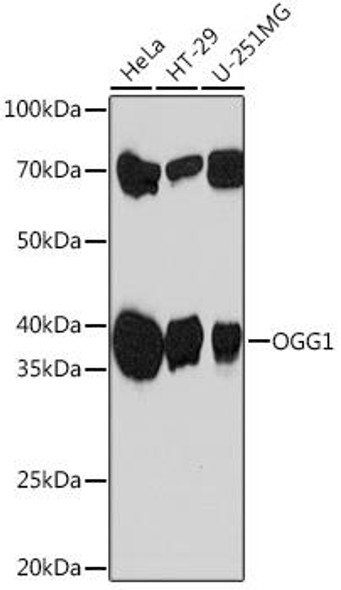
| Observed MW: | 36kDa |
| Synonyms: | OGG1, HMMH, HOGG1, MUTM, OGH1 |
| Background: | This gene encodes the enzyme responsible for the excision of 8-oxoguanine, a mutagenic base byproduct which occurs as a result of exposure to reactive oxygen. The action of this enzyme includes lyase activity for chain cleavage. Alternative splicing of the C-terminal region of this gene classifies splice variants into two major groups, type 1 and type 2, depending on the last exon of the sequence. Type 1 alternative splice variants end with exon 7 and type 2 end with exon 8. All variants share the N-terminal region in common, which contains a mitochondrial targeting signal that is essential for mitochondrial localization. Many alternative splice variants for this gene have been described, but the full-length nature for every variant has not been determined. |
| UniProt Protein Function: | OGG1: DNA repair enzyme that incises DNA at 8-oxoG residues. Excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N- methylformamidopyrimidine (FAPY) from damaged DNA. Has a beta- lyase activity that nicks DNA 3' to the lesion. Defects in OGG1 may be a cause of renal cell carcinoma (RCC). It is a heterogeneous group of sporadic or hereditary carcinoma derived from cells of the proximal renal tubular epithelium. It is subclassified into clear cell renal carcinoma (non-papillary carcinoma), papillary renal cell carcinoma, chromophobe renal cell carcinoma, collecting duct carcinoma with medullary carcinoma of the kidney, and unclassified renal cell carcinoma. Belongs to the type-1 OGG1 family. 8 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:EC 4.2.99.18; Lyase; DNA repair, damage; Deoxyribonuclease Chromosomal Location of Human Ortholog: 3p26.2 Cellular Component: nucleoplasm; nuclear matrix; mitochondrion; nuclear speck Molecular Function:protein binding; microtubule binding; endonuclease activity; DNA N-glycosylase activity; oxidized purine base lesion DNA N-glycosylase activity; damaged DNA binding; 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity Biological Process: response to drug; depurination; DNA repair; DNA catabolic process, endonucleolytic; response to estradiol stimulus; response to radiation; response to ethanol; base-excision repair, AP site formation; regulation of transcription, DNA-dependent; nucleotide-excision repair; base-excision repair; response to folic acid; regulation of protein import into nucleus, translocation; response to oxidative stress; acute inflammatory response; negative regulation of apoptosis; aging Disease: Renal Cell Carcinoma, Nonpapillary |
| NCBI Summary: | This gene encodes the enzyme responsible for the excision of 8-oxoguanine, a mutagenic base byproduct which occurs as a result of exposure to reactive oxygen. The action of this enzyme includes lyase activity for chain cleavage. Alternative splicing of the C-terminal region of this gene classifies splice variants into two major groups, type 1 and type 2, depending on the last exon of the sequence. Type 1 alternative splice variants end with exon 7 and type 2 end with exon 8. All variants share the N-terminal region in common, which contains a mitochondrial targeting signal that is essential for mitochondrial localization. Many alternative splice variants for this gene have been described, but the full-length nature for every variant has not been determined. [provided by RefSeq, Aug 2008] |
| UniProt Code: | O15527 |
| NCBI GenInfo Identifier: | 12643548 |
| NCBI Gene ID: | 4968 |
| NCBI Accession: | O15527.2 |
| UniProt Secondary Accession: | O15527,O00390, O00670, O00705, O14876, O95488, P78554 Q9BW42, Q9UIK0, Q9UIK1, Q9UIK2, A8K1E3, |
| UniProt Related Accession: | O15527 |
| Molecular Weight: | 345 |
| NCBI Full Name: | N-glycosylase/DNA lyase |
| NCBI Synonym Full Names: | 8-oxoguanine DNA glycosylase |
| NCBI Official Symbol: | OGG1 |
| NCBI Official Synonym Symbols: | HMMH; MUTM; OGH1; HOGG1 |
| NCBI Protein Information: | N-glycosylase/DNA lyase; AP lyase; OGG1 type 1f; 8-hydroxyguanine DNA glycosylase; DNA-apurinic or apyrimidinic site lyase |
| UniProt Protein Name: | N-glycosylase/DNA lyase |
| Protein Family: | N-glycosylase/DNA lyase |
| UniProt Gene Name: | OGG1 |
| UniProt Entry Name: | OGG1_HUMAN |