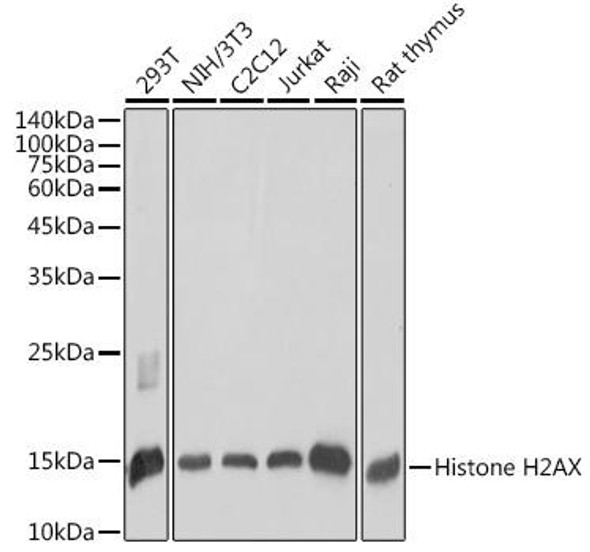
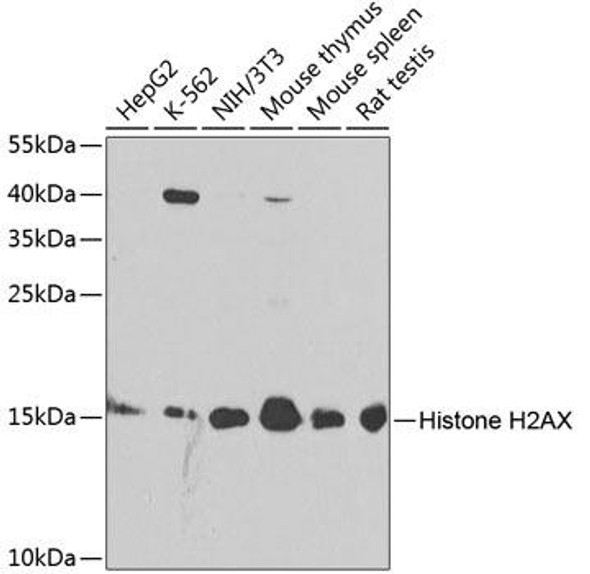
Cell Cycle Antibodies 1
Anti-Histone H2AX Antibody (CAB11540)
- SKU:
- CAB11540
- Product Type:
- Antibody
- Reactivity:
- Human
- Reactivity:
- Mouse
- Reactivity:
- Rat
- Host Species:
- Rabbit
- Isotype:
- IgG
- Antibody Type:
- Polyclonal Antibody
- Research Area:
- Cell Cycle
Description
| Antibody Name: | Anti-Histone H2AX Antibody |
| Antibody SKU: | CAB11540 |
| Antibody Size: | 20uL, 50uL, 100uL |
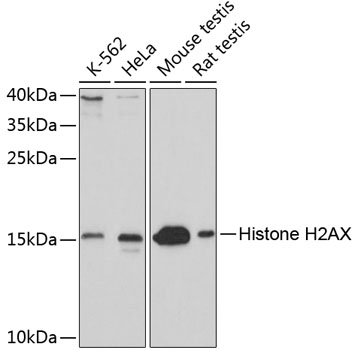
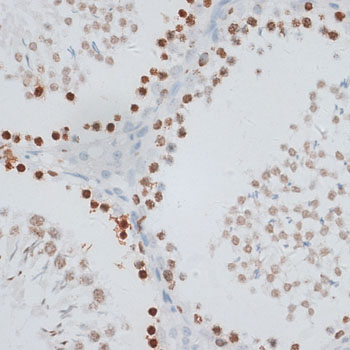
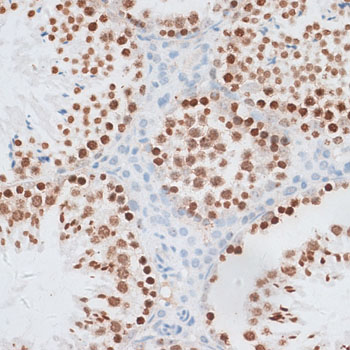
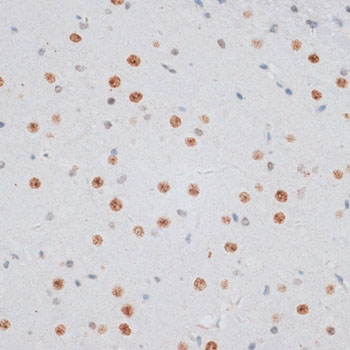
| Application: | WB IHC IF |
| Reactivity: | Human, Mouse, Rat, Other (Wide Range) |
| Host Species: | Rabbit |
| Immunogen: | Recombinant protein of human Histone H2AX. |
| Application: | WB IHC IF |
| Recommended Dilution: | WB 1:500 - 1:1000 IHC 1:50 - 1:200 IF 1:50 - 1:200 |
| Reactivity: | Human, Mouse, Rat, Other (Wide Range) |
| Positive Samples: | K-562, HeLa, Mouse testis, Rat testis |
| Immunogen: | Recombinant protein of human Histone H2AX. |
| Purification Method: | Affinity purification |
| Storage Buffer: | Store at -20'C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | Email for sequence |
| Gene ID: | 3014 |
| Uniprot: | P16104 |
| Cellular Location: | Chromosome, Nucleus |
| Calculated MW: | 15kDa |
| Observed MW: | 15kDa |
| Synonyms: | H2A.X, H2A/X, H2AX, Histone H2AX, H2AFX, histone H2AX, gamma H2A.X, GammaH2AX |
| Background: | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene encodes a replication-independent histone that is a member of the histone H2A family, and generates two transcripts through the use of the conserved stem-loop termination motif, and the polyA addition motif. |
| UniProt Protein Function: | H2AX: a histone that replaces conventional H2A in a subset of nucleosomes. Required for checkpoint-mediated arrest of cell cycle progression in response to low doses of ionizing radiation and for efficient repair of DNA double strand breaks (DSBs) specifically when modified by C-terminal phosphorylation. Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. Phosphorylated on S139 by ATM and DNA-PK in response to DNA double strand breaks (DSBs) generated by exogenous genotoxic agents and by stalled replication forks, and may also occur during meiotic recombination events and immunoglobulin class switching in lymphocytes. Phosphorylation can extend up to several thousand nucleosomes from the actual site of the DSB and may mark the surrounding chromatin for recruitment of proteins required for DNA damage signaling and repair. Widespread phosphorylation may also serve to amplify the damage signal or aid repair of persistent lesions. Dephosphorylation of S139 by PP2A is required for DNA DSB repair. Apparently phosphorylated on Y143 by WSTF, determining the relative recruitment of either DNA repair or pro-apoptotic factors. H2AXpY142 favors the recruitment of pro-apoptotic factors APBB1 and JNK1. In contrast, dephosphorylation of pY143 by EYA phosphatases favors the recruitment of MDC1-containing DNA repair complexes to the tail of phosphorylated pS139. Monoubiquitination of K119 by RING1 and RNF2/RING2 complex gives a specific tag for epigenetic transcriptional repression. Following DNA double-strand breaks (DSBs), it is ubiquitinated through 'K63' linkages by the E2 ligase UBE2N and the E3 ligases RNF8 and RNF168, leading to the recruitment of repair proteins to sites of DNA damage. |
| UniProt Protein Details: | Protein type:DNA repair, damage; DNA-binding; Helicase Chromosomal Location of Human Ortholog: 11q23.3 Cellular Component: chromosome, telomeric region; condensed nuclear chromosome; male germ cell nucleus; nuclear chromatin; nucleoplasm; nucleosome; nucleus; replication fork; XY body Molecular Function:damaged DNA binding; DNA binding; enzyme binding; histone binding; protein binding; protein heterodimerization activity Biological Process: cerebral cortex development; chromatin silencing; DNA damage checkpoint; DNA repair; double-strand break repair; double-strand break repair via homologous recombination; double-strand break repair via nonhomologous end joining; meiotic cell cycle; nucleosome assembly; positive regulation of DNA repair; response to DNA damage stimulus; response to ionizing radiation; spermatogenesis; viral reproduction |
| NCBI Summary: | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene encodes a replication-independent histone that is a member of the histone H2A family, and generates two transcripts through the use of the conserved stem-loop termination motif, and the polyA addition motif. [provided by RefSeq, Oct 2015] |
| UniProt Code: | P16104 |
| NCBI GenInfo Identifier: | 121992 |
| NCBI Gene ID: | 3014 |
| NCBI Accession: | P16104.2 |
| UniProt Secondary Accession: | P16104,Q4ZGJ7, Q6IAS5, |
| UniProt Related Accession: | P16104 |
| Molecular Weight: | 15,145 Da |
| NCBI Full Name: | Histone H2AX |
| NCBI Synonym Full Names: | H2A histone family member X |
| NCBI Official Symbol: | H2AFX |
| NCBI Official Synonym Symbols: | H2AX; H2A.X; H2A/X |
| NCBI Protein Information: | histone H2AX |
| UniProt Protein Name: | Histone H2AX |
| UniProt Synonym Protein Names: | Histone H2A.X |
| UniProt Gene Name: | H2AFX |
| UniProt Entry Name: | H2AX_HUMAN |