Cell Biology Antibodies 9
Anti-Acetyl-Histone H4-K16 Antibody (CAB5280)
- SKU:
- CAB5280
- Product Type:
- Antibody
- Applications:
- WB
- Applications:
- IF
- Applications:
- IP
- Applications:
- ChIP
- Reactivity:
- Human
- Reactivity:
- Mouse
- Reactivity:
- Rat
- Host Species:
- Rabbit
- Isotype:
- IgG
- Research Area:
- Cell Biology
Description
| Antibody Name: | Anti-Acetyl-Histone H4-K16 Antibody |
| Antibody SKU: | CAB5280 |
| Antibody Size: | 20uL, 50uL, 100uL |
| Application: | WB IF IP ChIP |
| Reactivity: | Human, Mouse, Rat, Other (Wide Range) |
| Host Species: | Rabbit |
| Immunogen: | A synthetic acetylated peptide around K16 of human Histone H4 (NP_001029249.1). |
| Application: | WB IF IP ChIP |
| Recommended Dilution: | WB 1:500 - 1:2000 IF 1:50 - 1:200 ChIP 1:50 - 1:200 |
| Reactivity: | Human, Mouse, Rat, Other (Wide Range) |
| Positive Samples: | Hela TSA, HeLa, C2C12 TSA, C2C12 |
| Immunogen: | A synthetic acetylated peptide around K16 of human Histone H4 (NP_001029249.1). |
| Purification Method: | Affinity purification |
| Storage Buffer: | Store at -20'C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | GGAK R |
| Gene ID: | 8370 |
| Uniprot: | P62805 |
| Cellular Location: | Chromosome, Nucleus |
| Calculated MW: | 11kDa |
| Observed MW: | 13kDa |
| Synonyms: | FO108, H4, H4/n, H4F2, H4FN, HIST2H4, Histone H4, HIST1H4A, HIST2H4A |
| Background: | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. This structure consists of approximately 146 bp of DNA wrapped around a nucleosome, an octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H4 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in a histone cluster on chromosome 1. This gene is one of four histone genes in the cluster that are duplicated; this record represents the centromeric copy. |
| UniProt Protein Function: | Function: Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. |
| UniProt Protein Details: | Subunit structure: The nucleosome is a histone octamer containing two molecules each of H2A, H2B, H3 and H4 assembled in one H3-H4 heterotetramer and two H2A-H2B heterodimers. The octamer wraps approximately 147 bp of DNA. Subcellular location: Nucleus. Chromosome. Post-translational modification: Acetylation at Lys-6 (H4K5ac), Lys-9 (H4K8ac), Lys-13 (H4K12ac) and Lys-17 (H4K16ac) occurs in coding regions of the genome but not in heterochromatin.Citrullination at Arg-4 (H4R3ci) by PADI4 impairs methylation.Monomethylation and asymmetric dimethylation at Arg-4 (H4R3me1 and H4R3me2a, respectively) by PRMT1 favors acetylation at Lys-9 (H4K8ac) and Lys-13 (H4K12ac). Demethylation is performed by JMJD6. Symmetric dimethylation on Arg-4 (H4R3me2s) by the PRDM1/PRMT5 complex may play a crucial role in the germ-cell lineage.Monomethylated, dimethylated or trimethylated at Lys-21 (H4K20me1, H4K20me2, H4K20me3). Monomethylation is performed by SET8. Trimethylation is performed by SUV420H1 and SUV420H2 and induces gene silencing. Ref.19 Ref.20 Ref.21 Ref.23 Ref.24 Ref.28Phosphorylated by PAK2 at Ser-48 (H4S47ph). This phosphorylation increases the association of H3.3-H4 with the histone chaperone HIRA, thus promoting nucleosome assembly of H3.3-H4 and inhibiting nucleosome assembly of H3.1-H4. Ref.28 Ref.36Ubiquitinated by the CUL4-DDB-RBX1 complex in response to ultraviolet irradiation. This may weaken the interaction between histones and DNA and facilitate DNA accessibility to repair proteins. Monoubiquitinated at Lys-92 of histone H4 (H4K91ub1) in response to DNA damage. The exact role of H4K91ub1 in DNA damage response is still unclear but it may function as a licensing signal for additional histone H4 post-translational modifications such as H4 Lys-21 methylation (H4K20me). Ref.27 Ref.30Sumoylated, which is associated with transcriptional repression. Ref.22Crotonylation (Kcr) is specifically present in male germ cells and marks testis-specific genes in post-meiotic cells, including X-linked genes that escape sex chromosome inactivation in haploid cells. Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors. It is also associated with post-meiotically activated genes on autosomes. Ref.35 Sequence similarities: Belongs to the histone H4 family. Sequence caution: The sequence AAI28106.1 differs from that shown. Reason: Frameshift at position 3. |
| NCBI Summary: | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. This structure consists of approximately 146 bp of DNA wrapped around a nucleosome, an octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a member of the histone H4 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in a histone cluster on chromosome 1. This gene is one of four histone genes in the cluster that are duplicated; this record represents the telomeric copy. [provided by RefSeq, Jul 2008] |
| UniProt Code: | P62805 |
| NCBI GenInfo Identifier: | 77539758 |
| NCBI Gene ID: | 554313 |
| NCBI Accession: | NP_001029249.1 |
| UniProt Secondary Accession: | P62805,P02304, P02305, Q6DRA9, Q6FGB8, Q6NWP7, A2VCL0 |
| UniProt Related Accession: | P62805 |
| Molecular Weight: | 11,367 Da |
| NCBI Full Name: | histone H4 |
| NCBI Synonym Full Names: | histone cluster 2, H4b |
| NCBI Official Symbol: | HIST2H4BÂ Â |
| NCBI Official Synonym Symbols: | H4/o  |
| NCBI Protein Information: | histone H4; histone 2, H4b |
| UniProt Protein Name: | Histone H4 |
| Protein Family: | Histone |
| UniProt Gene Name: | HIST1H4AÂ Â |
| UniProt Entry Name: | H4_HUMAN |
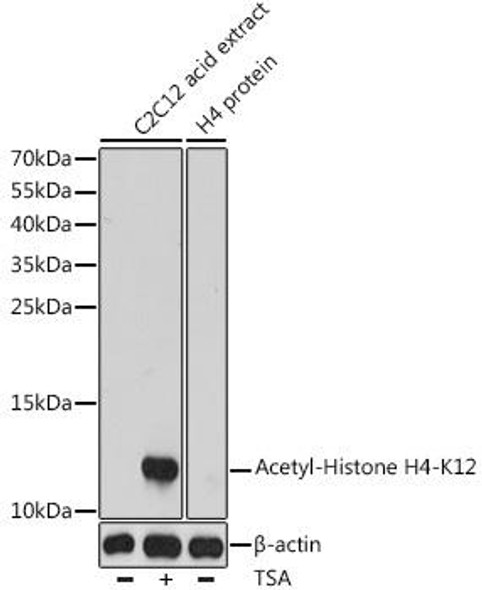
 | Western blot analysis of extracts of various cell lines, using Acetyl-Histone H4-K16 antibody at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) at 1:10000 dilution. Lysates/proteins: 25ug per lane. Blocking buffer: 3% nonfat dry milk in TBST. Detection: ECL Basic Kit. Exposure time: 90s. |
 | Immunofluorescence analysis of C6 cells treated by TSA (upper left) and untreated C6 cells (upper right) using Acetyl-Histone H4-K16 Rabbit pAb (red, A5280) at dilution of 1:100. Blue: DAPI for nuclear staining. |
 | Immunofluorescence analysis of C6 cells using Acetyl-Histone H4-K16 at dilution of 1:100. Blue: DAPI for nuclear staining. C6 cells were treated by TSA (1 uM) at 37°C for 18 hours. Blue: DAPI for nuclear staining. |
 | Immunofluorescence analysis of NIH/3T3 cells using Acetyl-Histone H4-K16 at dilution of 1:100. Blue: DAPI for nuclear staining. NIH/3T3 cells were treated by TSA (1 uM) at 37°C for 18 hours. Blue: DAPI for nuclear staining. |
 | Immunofluorescence analysis of U-2 OS cells using Acetyl-Histone H4-K16 at dilution of 1:100. Blue: DAPI for nuclear staining. U2OS cells were treated by TSA (1 uM) at 37°C for 18 hours. Blue: DAPI for nuclear staining. |