Description
| Antibody Name: | Acetyl-HIST1H2BC (K120) Antibody (PACO61285) |
| Antibody SKU: | PACO61285 |
| Size: | 50ul |
| Host Species: | Rabbit |
| Tested Applications: | ELISA, ICC, IF, ChIP |
| Recommended Dilutions: | ELISA:1:2000-1:10000, ICC:1:1-1:10, IF:1:1-1:10 |
| Species Reactivity: | Human |
| Immunogen: | Peptide sequence around site of Acetyl-Lys (120) derived from Human Histone H2B type 1-C/E/F/G/I |
| Form: | Liquid |
| Storage Buffer: | Preservative: 0.03% Proclin 300 Constituents: 50% Glycerol, 0.01M PBS, pH 7.4 |
| Purification Method: | Antigen Affinity Purified |
| Clonality: | Polyclonal |
| Isotype: | IgG |
| Conjugate: | Non-conjugated |
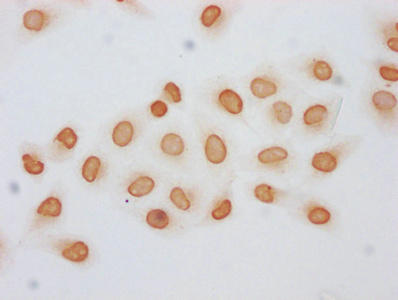 | Immunocytochemistry analysis of PACO61285 diluted at 1:5 and staining in Hela cells (treated with 30mM sodium butyrate for 4h) performed on a Leica BondTM system. The cells were fixed in 4% formaldehyde, permeabilized using 0.2% Triton X-100 and blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system. |
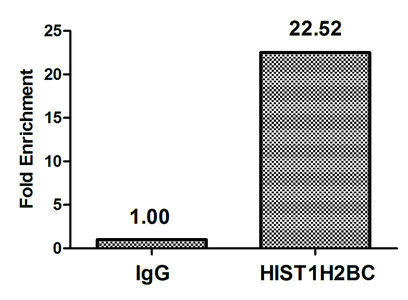 | Chromatin Immunoprecipitation Hela (106, treated with 30mM sodium butyrate for 4h) were treated with Micrococcal Nuclease, sonicated, and immunoprecipitated with 5µg anti-HIST1H2BC (PACO61285) or a control normal rabbit IgG. The resulting ChIP DNA was quantified using real-time PCR with primers against the beta -Globin promoter. |
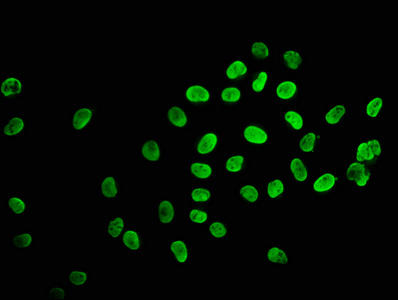 | Immunofluorescence staining of Hela cells (treated with 30mM sodium butyrate for 4h) with PACO61285 at 1:2.5, counter-stained with DAPI. The cells were fixed in 4% formaldehyde, permeabilized using 0.2% Triton X-100 and blocked in 10% normal Goat Serum. The cells were then incubated with the antibody overnight at 4°C. The secondary antibody was Alexa Fluor 488-congugated AffiniPure Goat Anti-Rabbit IgG(H+L). |
| Background: | Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. |
| Synonyms: | Histone H2B type 1-C/E/F/G/I (Histone H2B.1 A) (Histone H2B.a) (H2B/a) (Histone H2B.g) (H2B/g) (Histone H2B.h) (H2B/h) (Histone H2B.k) (H2B/k) (Histone H2B.l) (H2B/l), HIST1H2BC; HIST1H2BE; HIST1H2BF; HIST1H2BG; HIST1H2BI, H2BFL; H2BFH; H2BFG; H2BFA; H2BFK |
| UniProt Protein Function: | H2B1C: a core component of the nucleoosome. The nucleosome, a basic organizational unit of chromosomal DNA, is octrameric, consisting of two molecules each of histones H2B, H2A, H3, H4. The octamer wraps approximately 147 bp of DNA. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. |
| UniProt Protein Details: | Protein type:DNA-binding Chromosomal Location of Human Ortholog: 6p22.1 Cellular Component: cytoplasm; extracellular space; nucleoplasm; nucleus Molecular Function:DNA binding; protein binding Biological Process: antibacterial humoral response; defense response to Gram-positive bacterium; innate immune response in mucosa; nucleosome assembly |
| NCBI Summary: | Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. The protein has antibacterial and antifungal antimicrobial activity. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H2B family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6p22-p21.3. [provided by RefSeq, Aug 2015] |
| UniProt Code: | P62807 |
| NCBI GenInfo Identifier: | 290457686 |
| NCBI Gene ID: | 8339 |
| NCBI Accession: | P62807.4 |
| UniProt Secondary Accession: | P62807,P02278, Q3B872, Q4VB69, Q93078, Q93080, |
| UniProt Related Accession: | P62807 |
| Molecular Weight: | 13,906 Da |
| NCBI Full Name: | Histone H2B type 1-C/E/F/G/I |
| NCBI Synonym Full Names: | histone cluster 1, H2bg |
| NCBI Official Symbol: | HIST1H2BG |
| NCBI Official Synonym Symbols: | H2B/a; H2BFA; H2B.1A; dJ221C16.8 |
| NCBI Protein Information: | histone H2B type 1-C/E/F/G/I |
| UniProt Protein Name: | Histone H2B type 1-C/E/F/G/I |
| UniProt Synonym Protein Names: | Histone H2B.1 A; Histone H2B.a; H2B/a; Histone H2B.g; H2B/g; Histone H2B.h; H2B/h; Histone H2B.k; H2B/k; Histone H2B.l; H2B/l |
| Protein Family: | Histone |
| UniProt Gene Name: | HIST1H2BC |
| UniProt Entry Name: | H2B1C_HUMAN |






